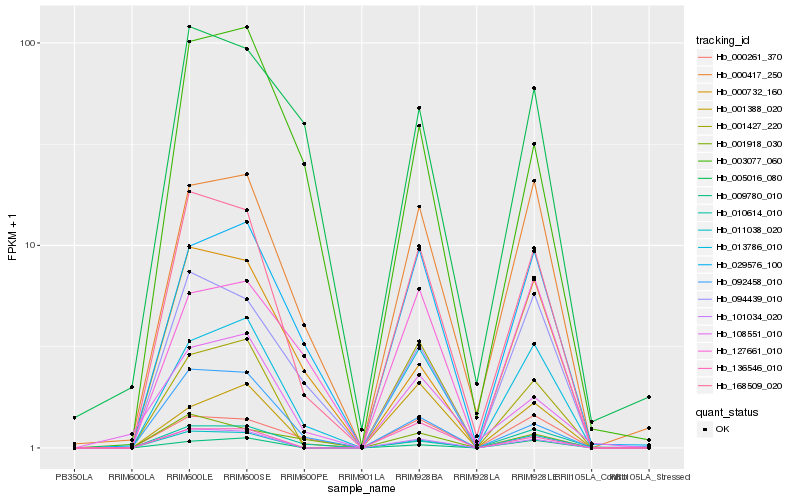
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168509_020 |
0.0 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |
| 2 |
Hb_101034_020 |
0.1059252097 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 3 |
Hb_013786_010 |
0.1241777439 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_011038_020 |
0.1248049437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000417_250 |
0.1387586486 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 6 |
Hb_094439_010 |
0.148409955 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 7 |
Hb_029576_100 |
0.1509221815 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 8 |
Hb_092458_010 |
0.1602580791 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_001918_030 |
0.1666625617 |
- |
- |
- |
| 10 |
Hb_000261_370 |
0.1692549349 |
- |
- |
- |
| 11 |
Hb_001427_220 |
0.1723248107 |
- |
- |
hypothetical protein CISIN_1g033375mg [Citrus sinensis] |
| 12 |
Hb_108551_010 |
0.1733727593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010614_010 |
0.1754144181 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 14 |
Hb_000732_160 |
0.1826857482 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 15 |
Hb_003077_060 |
0.1841946315 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 16 |
Hb_005016_080 |
0.1857631913 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 17 |
Hb_009780_010 |
0.1912135538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_136546_010 |
0.1941055989 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 19 |
Hb_127661_010 |
0.1972279904 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_001388_020 |
0.1973717971 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP20 [Jatropha curcas] |