| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
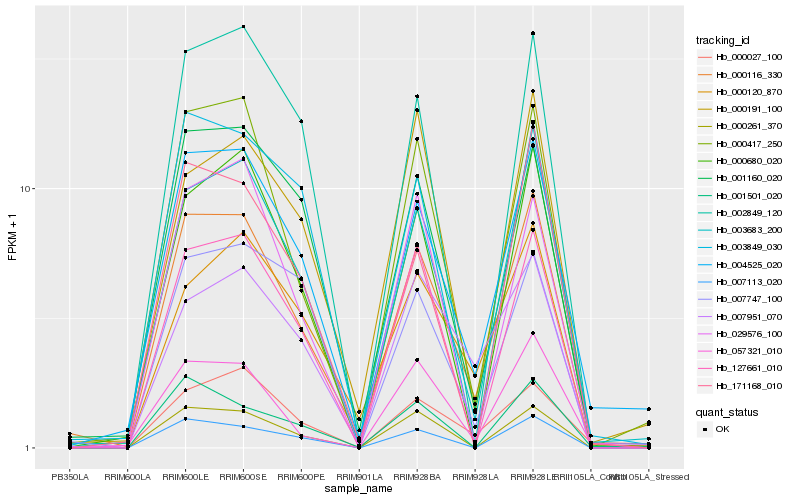
Hb_000116_330 |
0.0 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 2 |
Hb_003683_200 |
0.0948109776 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 3 |
Hb_004525_020 |
0.0967690066 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 4 |
Hb_000417_250 |
0.1051573314 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 5 |
Hb_001160_020 |
0.1281722902 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 6 |
Hb_000680_020 |
0.1310478044 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000261_370 |
0.1321849507 |
- |
- |
- |
| 8 |
Hb_007113_020 |
0.1323789298 |
- |
- |
- |
| 9 |
Hb_127661_010 |
0.1359764951 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_057321_010 |
0.1393803726 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_002849_120 |
0.1403507844 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 12 |
Hb_000120_870 |
0.1435215642 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 13 |
Hb_003849_030 |
0.1449113388 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 14 |
Hb_029576_100 |
0.1464213876 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 15 |
Hb_007951_070 |
0.1469099023 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_171168_010 |
0.1470513794 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000027_100 |
0.1501088737 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 18 |
Hb_000191_100 |
0.1507237895 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_001501_020 |
0.1520839083 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 20 |
Hb_007747_100 |
0.157082055 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |