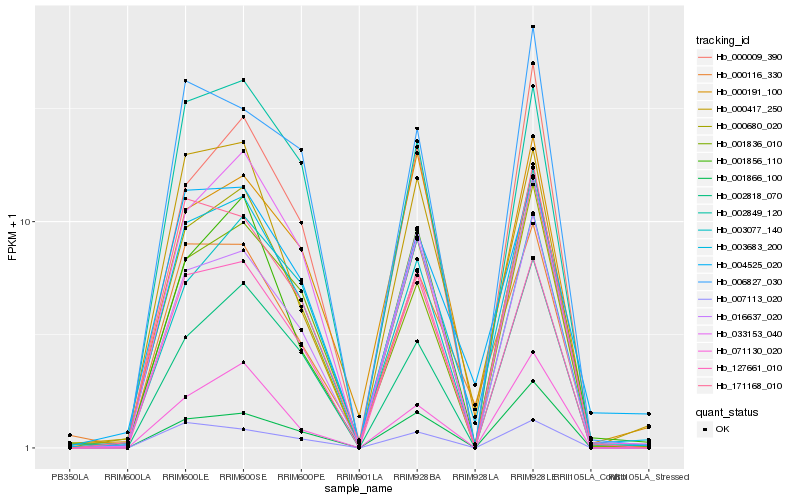
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003683_200 |
0.0 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 2 |
Hb_000680_020 |
0.0763874273 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000116_330 |
0.0948109776 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 4 |
Hb_001836_010 |
0.1019216391 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 5 |
Hb_001856_110 |
0.1057612395 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 6 |
Hb_004525_020 |
0.1064964907 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 7 |
Hb_002849_120 |
0.1103896862 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 8 |
Hb_002818_070 |
0.112098994 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_071130_020 |
0.1135524875 |
- |
- |
- |
| 10 |
Hb_000009_390 |
0.1144590404 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_171168_010 |
0.1152655603 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000417_250 |
0.115306988 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 13 |
Hb_127661_010 |
0.119104997 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001866_100 |
0.1205762122 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 15 |
Hb_003077_140 |
0.1215274126 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 16 |
Hb_007113_020 |
0.1224246483 |
- |
- |
- |
| 17 |
Hb_006827_030 |
0.1239457811 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 18 |
Hb_033153_040 |
0.1270319981 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000191_100 |
0.127799615 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_016637_020 |
0.1303955076 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |