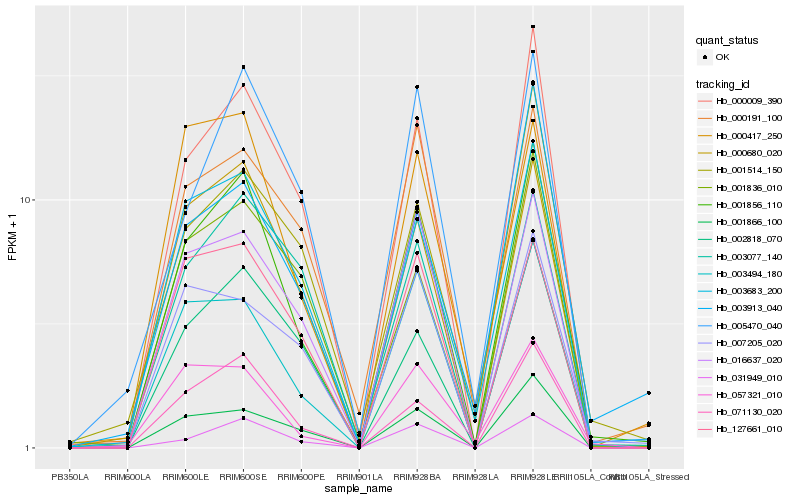
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_110 |
0.0 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 2 |
Hb_000009_390 |
0.0927929655 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_071130_020 |
0.0983203968 |
- |
- |
- |
| 4 |
Hb_003683_200 |
0.1057612395 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 5 |
Hb_031949_010 |
0.1080743496 |
- |
- |
- |
| 6 |
Hb_000680_020 |
0.1094869579 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_003494_180 |
0.1178089103 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 8 |
Hb_001866_100 |
0.1234156465 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 9 |
Hb_016637_020 |
0.1293142247 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 10 |
Hb_002818_070 |
0.1382643602 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_005470_040 |
0.1402630084 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 12 |
Hb_000191_100 |
0.1429571323 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_057321_010 |
0.143211968 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_001514_150 |
0.1467113145 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 15 |
Hb_001836_010 |
0.1472672843 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 16 |
Hb_003913_040 |
0.1506263971 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 17 |
Hb_003077_140 |
0.1508683137 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 18 |
Hb_127661_010 |
0.1510318529 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000417_250 |
0.1511023587 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 20 |
Hb_007205_020 |
0.1525232201 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |