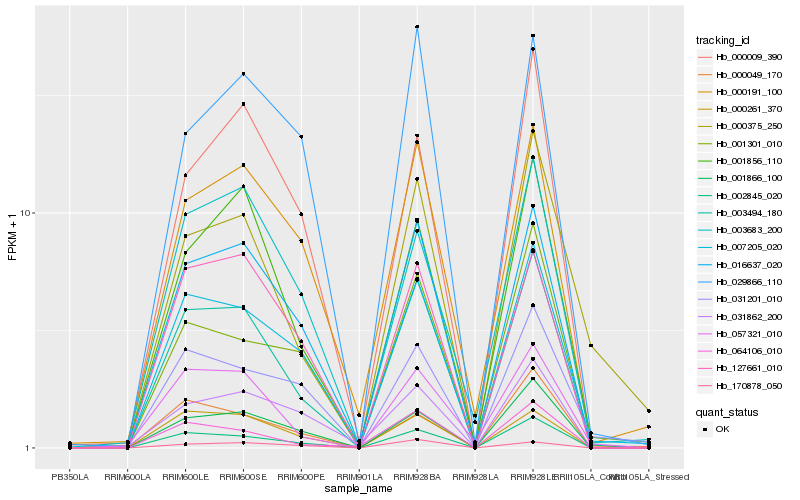
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003494_180 |
0.0 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 2 |
Hb_002845_020 |
0.0764049718 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 3 |
Hb_016637_020 |
0.087612122 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 4 |
Hb_007205_020 |
0.0957708111 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 5 |
Hb_001866_100 |
0.1098594013 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 6 |
Hb_057321_010 |
0.1176143082 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 7 |
Hb_001856_110 |
0.1178089103 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 8 |
Hb_064106_010 |
0.118597954 |
- |
- |
PREDICTED: uncharacterized protein LOC104878249 [Vitis vinifera] |
| 9 |
Hb_000261_370 |
0.1273393116 |
- |
- |
- |
| 10 |
Hb_127661_010 |
0.1323234492 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000191_100 |
0.1354760516 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_000049_170 |
0.1395381584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000375_250 |
0.1421707551 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 14 |
Hb_000009_390 |
0.1427073357 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001301_010 |
0.1434384416 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_031862_200 |
0.146665323 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 17 |
Hb_003683_200 |
0.1479514448 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 18 |
Hb_031201_010 |
0.1489419365 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_029866_110 |
0.1513150814 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 20 |
Hb_170878_050 |
0.1531577287 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |