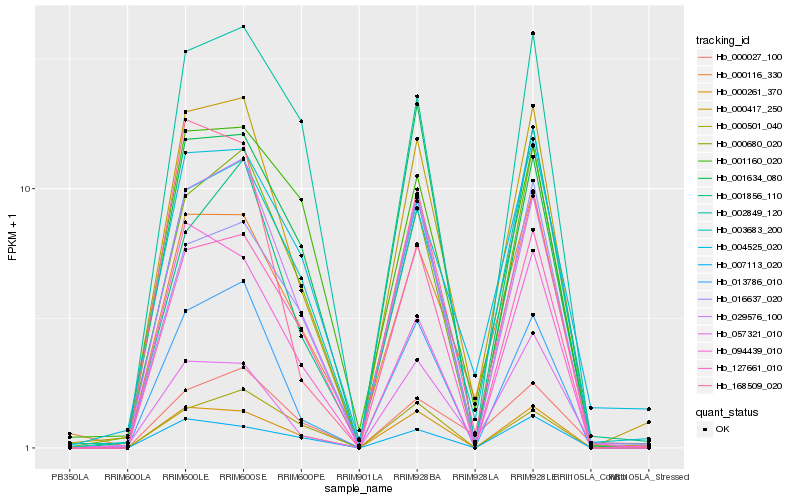
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_250 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 2 |
Hb_029576_100 |
0.085158699 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 3 |
Hb_013786_010 |
0.0963966718 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000116_330 |
0.1051573314 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_000261_370 |
0.1059810534 |
- |
- |
- |
| 6 |
Hb_000680_020 |
0.1121239569 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_127661_010 |
0.1146932314 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_003683_200 |
0.115306988 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_004525_020 |
0.125573845 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 10 |
Hb_057321_010 |
0.1297147336 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_094439_010 |
0.1367130939 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 12 |
Hb_168509_020 |
0.1387586486 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |
| 13 |
Hb_002849_120 |
0.1426957517 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 14 |
Hb_007113_020 |
0.1430873328 |
- |
- |
- |
| 15 |
Hb_001160_020 |
0.1504833452 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 16 |
Hb_001856_110 |
0.1511023587 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 17 |
Hb_001634_080 |
0.1564017626 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_016637_020 |
0.1575599068 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 19 |
Hb_000027_100 |
0.1575978047 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 20 |
Hb_000501_040 |
0.1588427692 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |