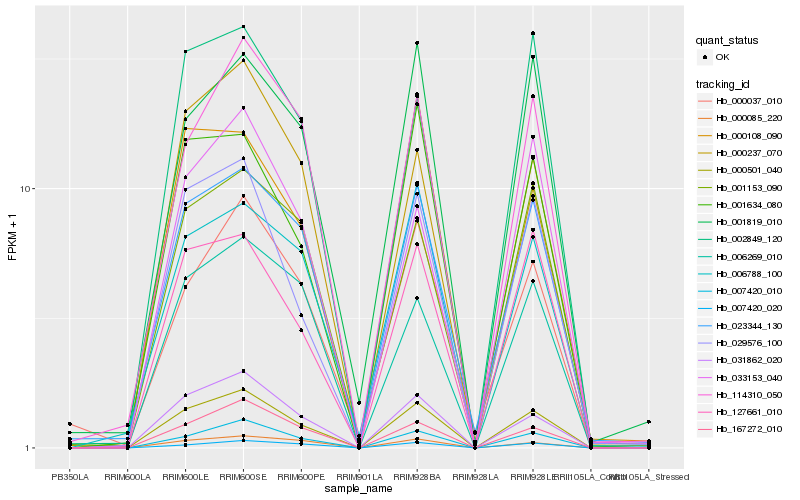
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000501_040 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_007420_010 |
0.0700887845 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 3 |
Hb_031862_020 |
0.0741413121 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 4 |
Hb_167272_010 |
0.0917289753 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 5 |
Hb_029576_100 |
0.0922501815 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 6 |
Hb_127661_010 |
0.0948593705 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_023344_130 |
0.0990314698 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_007420_020 |
0.1028885833 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_000085_220 |
0.1068684162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001634_080 |
0.1080648029 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_114310_050 |
0.109807933 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 12 |
Hb_006269_010 |
0.1131942171 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 13 |
Hb_001153_090 |
0.1153598871 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 14 |
Hb_006788_100 |
0.1177330663 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002849_120 |
0.1177970881 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 16 |
Hb_000108_090 |
0.1189368985 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 17 |
Hb_001819_010 |
0.1239369899 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 18 |
Hb_033153_040 |
0.1253100404 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000237_070 |
0.1271037641 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 20 |
Hb_000037_010 |
0.127324156 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |