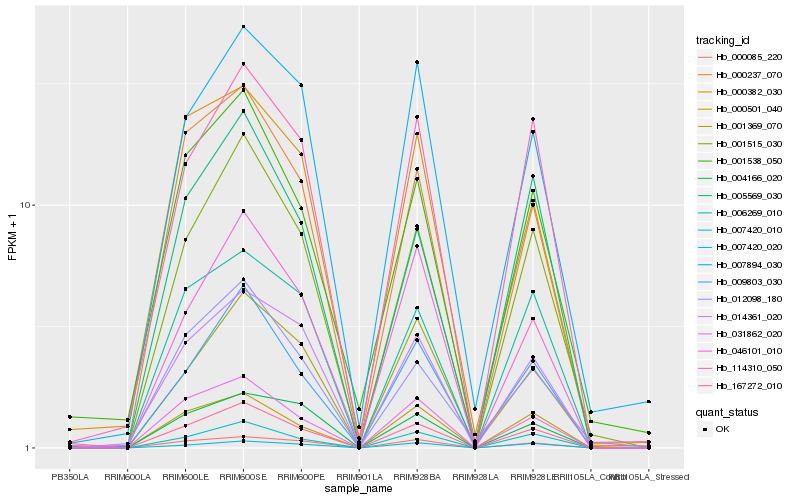
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167272_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 2 |
Hb_001515_030 |
0.0626205885 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_007420_010 |
0.0686576395 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 4 |
Hb_031862_020 |
0.0701833893 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 5 |
Hb_000237_070 |
0.0816028277 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 6 |
Hb_009803_030 |
0.0851997021 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 7 |
Hb_000085_220 |
0.0859574199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_014361_020 |
0.0913349612 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 9 |
Hb_000501_040 |
0.0917289753 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_114310_050 |
0.0942774627 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 11 |
Hb_006269_010 |
0.0963361458 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 12 |
Hb_005569_030 |
0.1016724392 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 13 |
Hb_046101_010 |
0.1046103895 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 14 |
Hb_007420_020 |
0.105994041 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 15 |
Hb_001369_070 |
0.1090480005 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_004166_020 |
0.1105623835 |
- |
- |
- |
| 17 |
Hb_001538_050 |
0.1145272362 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 18 |
Hb_007894_030 |
0.1151069502 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000382_030 |
0.1167800143 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 20 |
Hb_012098_180 |
0.120386382 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |