| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
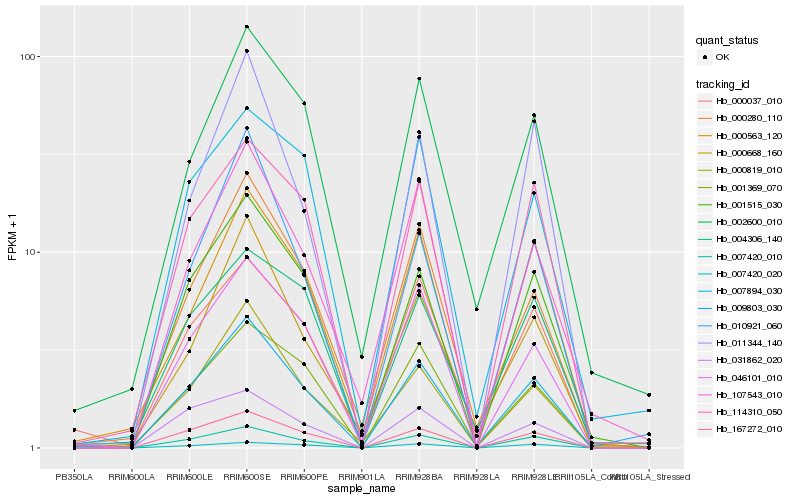
Hb_009803_030 |
0.0 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 2 |
Hb_007420_010 |
0.0800237157 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 3 |
Hb_167272_010 |
0.0851997021 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 4 |
Hb_000819_010 |
0.0858279259 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 5 |
Hb_001515_030 |
0.087982067 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000280_110 |
0.0896689787 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 7 |
Hb_046101_010 |
0.0928277927 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 8 |
Hb_000037_010 |
0.099662131 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 9 |
Hb_114310_050 |
0.1039140825 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 10 |
Hb_001369_070 |
0.1088968942 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000668_160 |
0.1109064244 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 12 |
Hb_000563_120 |
0.1116380485 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 13 |
Hb_002600_010 |
0.114006918 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_107543_010 |
0.118337866 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 15 |
Hb_010921_060 |
0.1225244611 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_011344_140 |
0.1227632337 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_031862_020 |
0.1241591192 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 18 |
Hb_007894_030 |
0.1242379022 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 19 |
Hb_004306_140 |
0.1249960009 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 20 |
Hb_007420_020 |
0.1256702254 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |