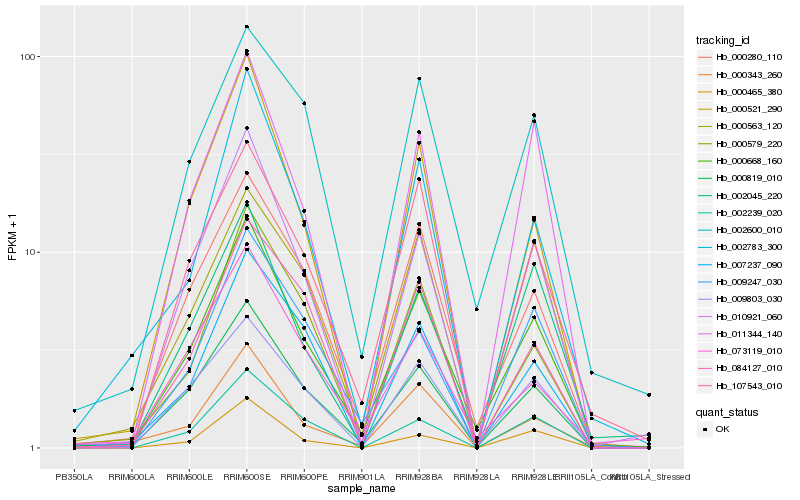
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_160 |
0.0 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 2 |
Hb_000819_010 |
0.0826219274 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 3 |
Hb_000280_110 |
0.0935406936 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 4 |
Hb_010921_060 |
0.0986860394 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_009803_030 |
0.1109064244 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 6 |
Hb_002600_010 |
0.117763527 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_107543_010 |
0.1234460791 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 8 |
Hb_002239_020 |
0.1252089283 |
- |
- |
MLO, putative [Theobroma cacao] |
| 9 |
Hb_011344_140 |
0.126004727 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 10 |
Hb_000521_290 |
0.1261522547 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 11 |
Hb_002783_300 |
0.1279578716 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000343_260 |
0.1286557846 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 13 |
Hb_002045_220 |
0.13027982 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009247_030 |
0.132910339 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 15 |
Hb_073119_010 |
0.1348524521 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 16 |
Hb_000579_220 |
0.1378008086 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_007237_090 |
0.1391686031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000563_120 |
0.1440842413 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 19 |
Hb_000465_380 |
0.1447418531 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_084127_010 |
0.1451030923 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |