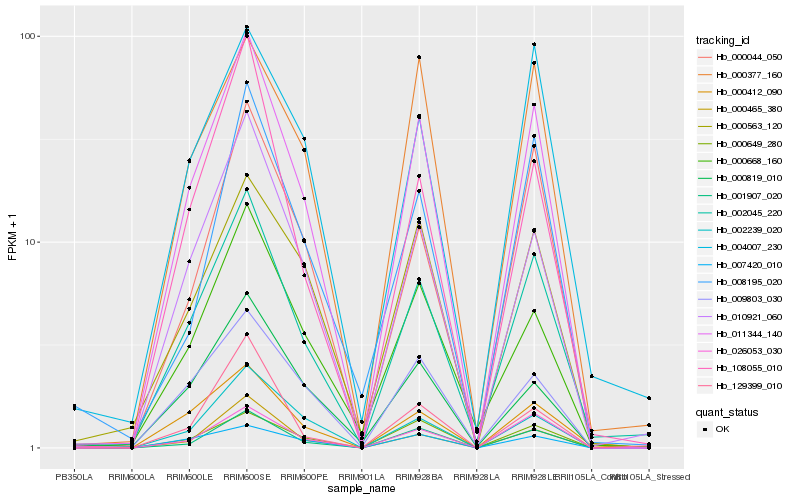
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_140 |
0.0 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_002045_220 |
0.0809940105 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001907_020 |
0.0814538524 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 4 |
Hb_010921_060 |
0.1038604094 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000465_380 |
0.1074983936 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000412_090 |
0.110896416 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 7 |
Hb_000649_280 |
0.114158428 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 8 |
Hb_026053_030 |
0.1142998208 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 9 |
Hb_002239_020 |
0.1180153965 |
- |
- |
MLO, putative [Theobroma cacao] |
| 10 |
Hb_009803_030 |
0.1227632337 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 11 |
Hb_000668_160 |
0.126004727 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 12 |
Hb_000563_120 |
0.1266312168 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 13 |
Hb_007420_010 |
0.1266508052 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000044_050 |
0.1289555051 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000377_160 |
0.134300958 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 16 |
Hb_000819_010 |
0.1390361137 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 17 |
Hb_108055_010 |
0.1396351519 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_129399_010 |
0.1459245607 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_004007_230 |
0.1463762486 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008195_020 |
0.1487933167 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |