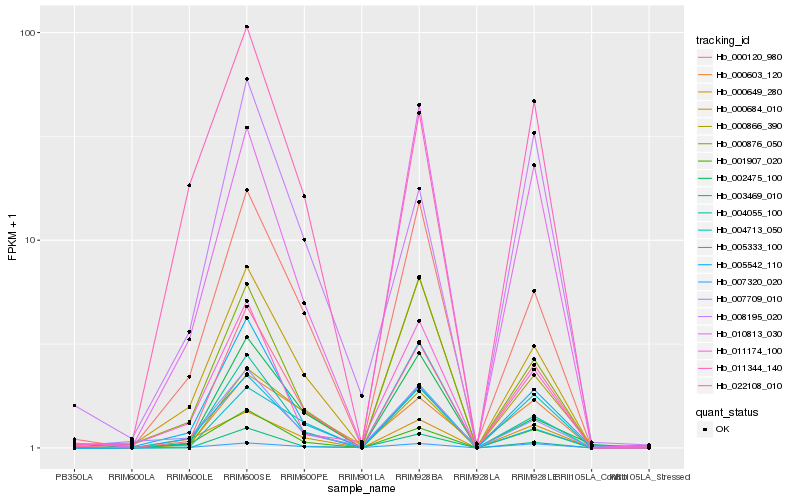
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022108_010 |
0.0 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Glycine soja] |
| 2 |
Hb_004055_100 |
0.0883843687 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 3 |
Hb_001907_020 |
0.1035646455 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 4 |
Hb_005542_110 |
0.1229272104 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 5 |
Hb_000684_010 |
0.1313170542 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000120_980 |
0.1328655917 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 7 |
Hb_003469_010 |
0.1486519129 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 8 |
Hb_004713_050 |
0.1527222397 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |
| 9 |
Hb_000876_050 |
0.1592577975 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 10 |
Hb_010813_030 |
0.1631945581 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 11 |
Hb_011174_100 |
0.1633791537 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_008195_020 |
0.1645734975 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 13 |
Hb_005333_100 |
0.1656337511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000649_280 |
0.169972095 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 15 |
Hb_002475_100 |
0.1709386863 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 16 |
Hb_011344_140 |
0.1737802176 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_007709_010 |
0.1752562564 |
- |
- |
- |
| 18 |
Hb_000866_390 |
0.1760767231 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 19 |
Hb_007320_020 |
0.1834979726 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 20 |
Hb_000603_120 |
0.1861274087 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |