| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
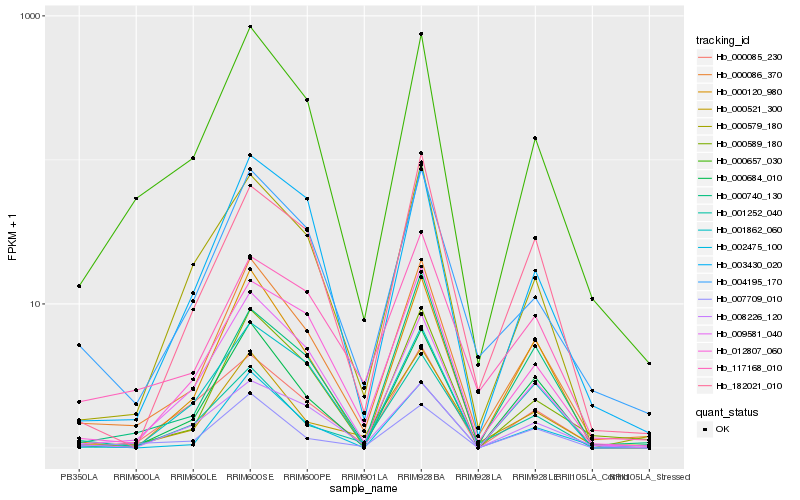
Hb_000086_370 |
0.0 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 2 |
Hb_000657_030 |
0.0910469248 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 3 |
Hb_000521_300 |
0.1084512795 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 4 |
Hb_004195_170 |
0.1107575263 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000120_980 |
0.1185575755 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 6 |
Hb_000589_180 |
0.1206835472 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 7 |
Hb_012807_060 |
0.1237097422 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000684_010 |
0.1263432947 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_001252_040 |
0.1272300227 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 10 |
Hb_000579_180 |
0.1281180548 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 11 |
Hb_009581_040 |
0.1284575973 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 12 |
Hb_003430_020 |
0.1348570529 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_002475_100 |
0.1377154755 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 14 |
Hb_182021_010 |
0.1378482998 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 15 |
Hb_001862_060 |
0.1396074592 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007709_010 |
0.1401300644 |
- |
- |
- |
| 17 |
Hb_000740_130 |
0.1407599867 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 18 |
Hb_000085_230 |
0.1417977428 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 19 |
Hb_117168_010 |
0.1419898233 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 20 |
Hb_008226_120 |
0.1450340652 |
- |
- |
ATP binding protein, putative [Ricinus communis] |