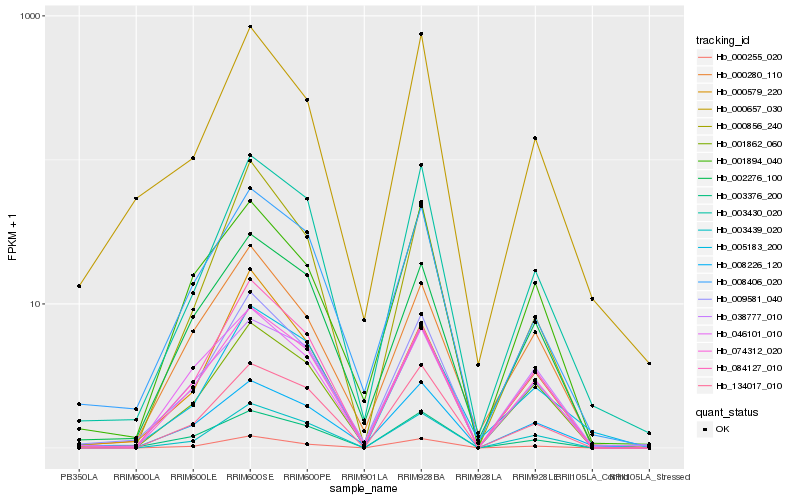
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009581_040 |
0.0 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 2 |
Hb_003430_020 |
0.0839609381 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_008406_020 |
0.0882268368 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 4 |
Hb_074312_020 |
0.089465722 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_001862_060 |
0.0895389575 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000255_020 |
0.0966106765 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 7 |
Hb_002276_100 |
0.1001411621 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 8 |
Hb_008226_120 |
0.1010722689 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_003439_020 |
0.1025692045 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1-like [Solanum lycopersicum] |
| 10 |
Hb_084127_010 |
0.1030556061 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000280_110 |
0.104555015 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 12 |
Hb_000657_030 |
0.1047958671 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 13 |
Hb_001894_040 |
0.1051266721 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003376_200 |
0.1079022926 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_000579_220 |
0.1089953696 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_005183_200 |
0.1186641095 |
- |
- |
EG964 [Manihot glaziovii] |
| 17 |
Hb_134017_010 |
0.1192226065 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_000856_240 |
0.119412871 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_046101_010 |
0.1212293608 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 20 |
Hb_038777_010 |
0.1218282252 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |