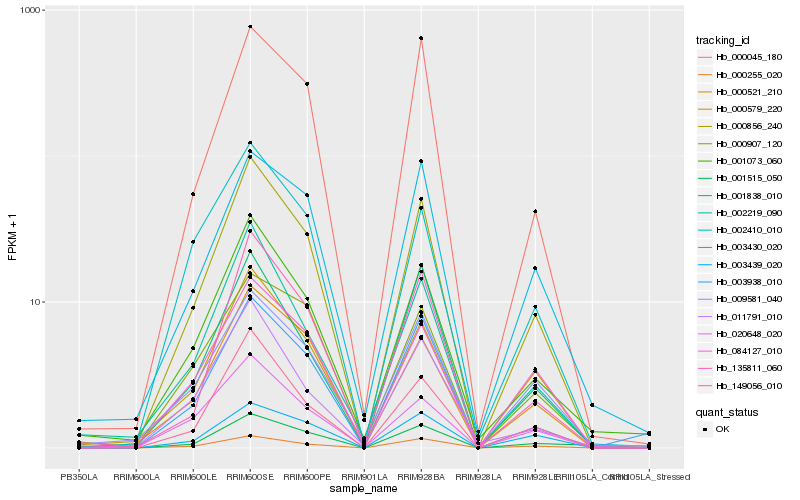
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_240 |
0.0 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_149056_010 |
0.082605513 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 3 |
Hb_000255_020 |
0.0893934502 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 4 |
Hb_000521_210 |
0.0897837568 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_001073_060 |
0.0945418625 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 6 |
Hb_000579_220 |
0.0971099585 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_135811_060 |
0.0993851193 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 8 |
Hb_000045_180 |
0.1046528856 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 9 |
Hb_003938_010 |
0.1101283644 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 10 |
Hb_084127_010 |
0.1107100375 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_020648_020 |
0.1116462841 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 12 |
Hb_002219_090 |
0.111771425 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 13 |
Hb_009581_040 |
0.119412871 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 14 |
Hb_001838_010 |
0.1201430441 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 15 |
Hb_011791_010 |
0.120208472 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 16 |
Hb_003439_020 |
0.1210561325 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1-like [Solanum lycopersicum] |
| 17 |
Hb_002410_010 |
0.1255972237 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 18 |
Hb_001515_050 |
0.130163669 |
- |
- |
PREDICTED: uncharacterized protein LOC105636626 [Jatropha curcas] |
| 19 |
Hb_000907_120 |
0.131096225 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription repressor KAN1-like [Populus euphratica] |
| 20 |
Hb_003430_020 |
0.1317945205 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |