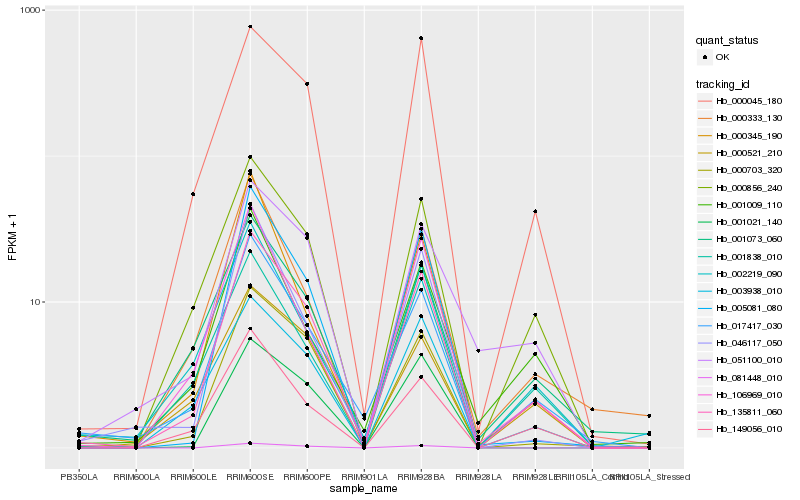
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135811_060 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 2 |
Hb_017417_030 |
0.0992359496 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 3 |
Hb_000856_240 |
0.0993851193 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000703_320 |
0.1055830219 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_000045_180 |
0.1118676369 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 6 |
Hb_149056_010 |
0.1190124764 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 7 |
Hb_005081_080 |
0.1238167437 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 8 |
Hb_001073_060 |
0.1267136466 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 9 |
Hb_003938_010 |
0.1325083405 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 10 |
Hb_002219_090 |
0.1341258828 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 11 |
Hb_106969_010 |
0.138210559 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 12 |
Hb_046117_050 |
0.1392064336 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 13 |
Hb_000333_130 |
0.1400725746 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_081448_010 |
0.1427798233 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 15 |
Hb_001021_140 |
0.143353126 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 16 |
Hb_051100_010 |
0.1437855242 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_000521_210 |
0.1445420669 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_001838_010 |
0.1456344137 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 19 |
Hb_001009_110 |
0.1461813852 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 20 |
Hb_000345_190 |
0.1483260761 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |