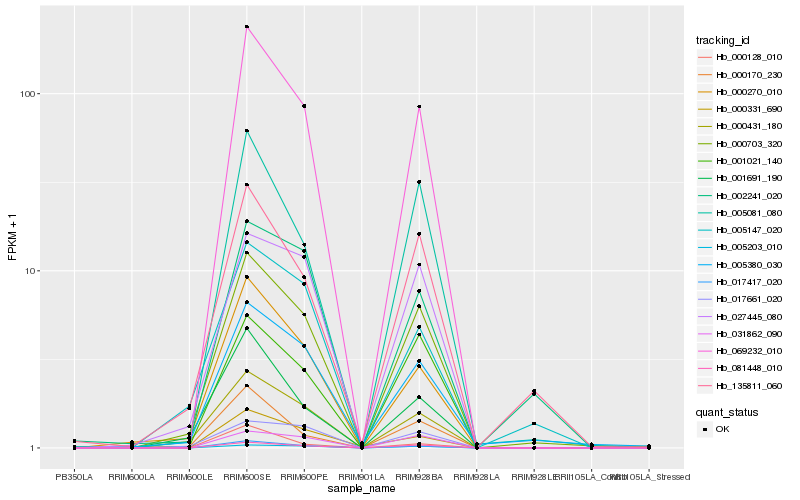
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069232_010 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Vitis vinifera] |
| 2 |
Hb_017417_020 |
0.0589127141 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_081448_010 |
0.061866811 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 4 |
Hb_000703_320 |
0.0985824513 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_000270_010 |
0.111501122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000331_690 |
0.1176706123 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 7 |
Hb_005203_010 |
0.1190432969 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 8 |
Hb_005081_080 |
0.1200305159 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 9 |
Hb_000431_180 |
0.1235359659 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 10 |
Hb_031862_090 |
0.137124282 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 11 |
Hb_001021_140 |
0.1400159071 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 12 |
Hb_005380_030 |
0.1423869228 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001691_190 |
0.1533438371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000170_230 |
0.1568665209 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 15 |
Hb_027445_080 |
0.1583674856 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 16 |
Hb_135811_060 |
0.1603397837 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 17 |
Hb_000128_010 |
0.1649099409 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 18 |
Hb_002241_020 |
0.1721072909 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 19 |
Hb_005147_020 |
0.1756393739 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 20 |
Hb_017661_020 |
0.1775855386 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |