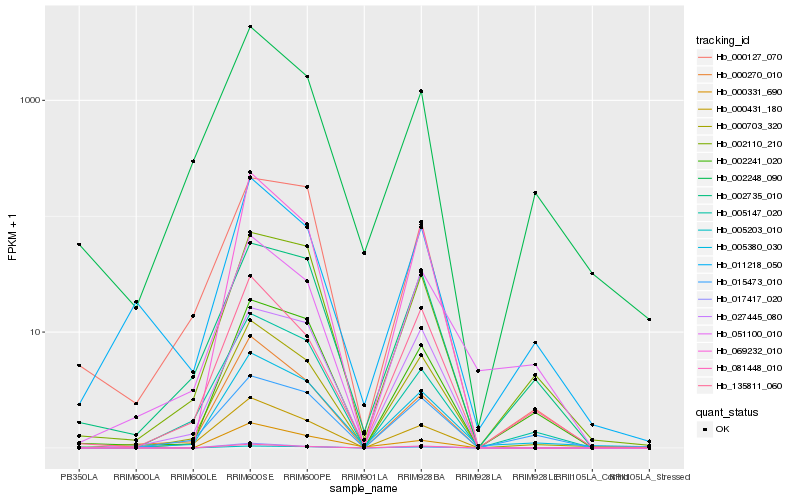
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005380_030 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000703_320 |
0.1024776415 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_002110_210 |
0.1090051593 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 4 |
Hb_002241_020 |
0.1221798636 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_005147_020 |
0.1309138134 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 6 |
Hb_002248_090 |
0.1338946231 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 7 |
Hb_002735_010 |
0.1339271241 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 8 |
Hb_000431_180 |
0.1395166048 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 9 |
Hb_069232_010 |
0.1423869228 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Vitis vinifera] |
| 10 |
Hb_027445_080 |
0.1464012169 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 11 |
Hb_015473_010 |
0.1472458462 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 12 |
Hb_000331_690 |
0.1485284982 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 13 |
Hb_000270_010 |
0.1489531346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_135811_060 |
0.1500779217 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 15 |
Hb_051100_010 |
0.1520582691 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_005203_010 |
0.1522114176 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 17 |
Hb_011218_050 |
0.1546520144 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_081448_010 |
0.1547669353 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 19 |
Hb_000127_070 |
0.155914403 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 20 |
Hb_017417_020 |
0.1585286365 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |