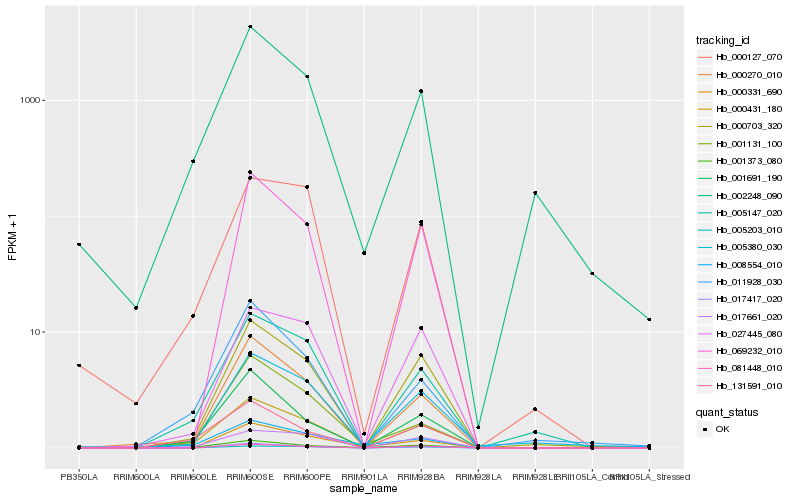
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_180 |
0.0 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 2 |
Hb_000703_320 |
0.0995365294 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_000270_010 |
0.1008987741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005147_020 |
0.1059598238 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 5 |
Hb_131591_010 |
0.1197479182 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 6 |
Hb_017661_020 |
0.122084739 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 7 |
Hb_069232_010 |
0.1235359659 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Vitis vinifera] |
| 8 |
Hb_001691_190 |
0.1276347865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_017417_020 |
0.1341365124 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 10 |
Hb_001373_080 |
0.1357704273 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005380_030 |
0.1395166048 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011928_030 |
0.1404770395 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 13 |
Hb_027445_080 |
0.1432377037 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 14 |
Hb_081448_010 |
0.14346901 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 15 |
Hb_000127_070 |
0.1498188506 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 16 |
Hb_002248_090 |
0.1507786548 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 17 |
Hb_008554_010 |
0.1543219619 |
- |
- |
- |
| 18 |
Hb_005203_010 |
0.1578401418 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 19 |
Hb_000331_690 |
0.1585463458 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 20 |
Hb_001131_100 |
0.159824049 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |