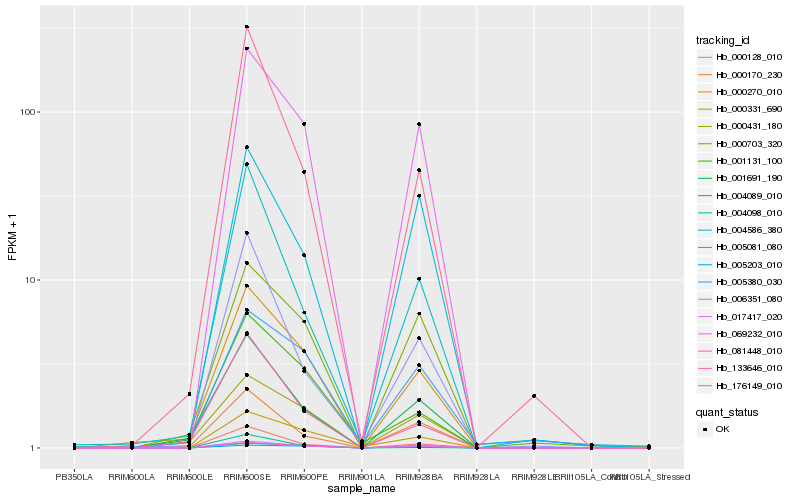
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017417_020 |
0.0 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_069232_010 |
0.0589127141 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Vitis vinifera] |
| 3 |
Hb_000270_010 |
0.0902395816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000331_690 |
0.1074625453 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 5 |
Hb_081448_010 |
0.1198050448 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 6 |
Hb_000128_010 |
0.1248941416 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_000431_180 |
0.1341365124 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 8 |
Hb_000703_320 |
0.1354737644 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_001691_190 |
0.140724544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004098_010 |
0.1452316174 |
- |
- |
- |
| 11 |
Hb_001131_100 |
0.1496578261 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 12 |
Hb_176149_010 |
0.1501980595 |
- |
- |
hypothetical protein JCGZ_16793 [Jatropha curcas] |
| 13 |
Hb_005081_080 |
0.1576931723 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 14 |
Hb_005380_030 |
0.1585286365 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004586_380 |
0.1594346198 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 16 |
Hb_004089_010 |
0.1611526557 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 17 |
Hb_000170_230 |
0.162622754 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006351_080 |
0.1643103849 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 19 |
Hb_005203_010 |
0.1665040636 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 20 |
Hb_133646_010 |
0.1697693048 |
- |
- |
hypothetical protein RCOM_1044030 [Ricinus communis] |