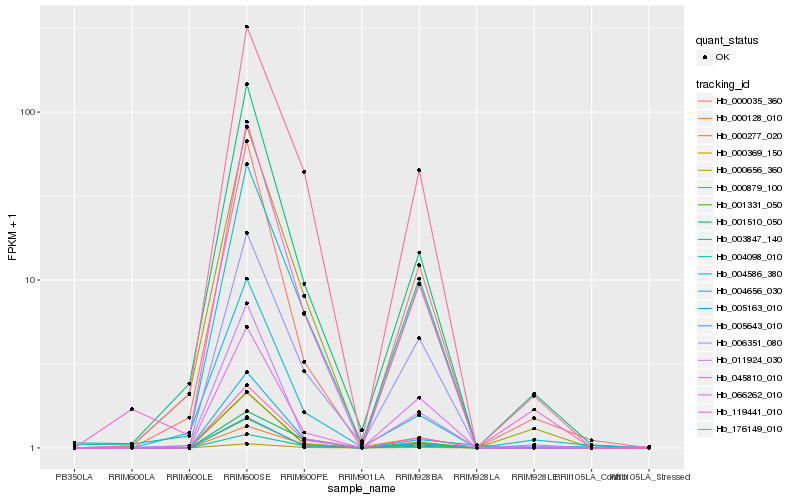
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119441_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0477971mg, partial [Citrus sinensis] |
| 2 |
Hb_000369_150 |
0.0239272987 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20185 [Jatropha curcas] |
| 3 |
Hb_005643_010 |
0.0505183645 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001331_050 |
0.0532543444 |
- |
- |
hypothetical protein VITISV_022439 [Vitis vinifera] |
| 5 |
Hb_000879_100 |
0.0600435343 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_004098_010 |
0.0629236021 |
- |
- |
- |
| 7 |
Hb_000128_010 |
0.0772937359 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_176149_010 |
0.0797575645 |
- |
- |
hypothetical protein JCGZ_16793 [Jatropha curcas] |
| 9 |
Hb_000656_360 |
0.0865267952 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 10 |
Hb_006351_080 |
0.0927363604 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 11 |
Hb_066262_010 |
0.0945664636 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 12 |
Hb_004586_380 |
0.1010337306 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 13 |
Hb_003847_140 |
0.1014916391 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005163_010 |
0.1111722147 |
- |
- |
PREDICTED: probable terpene synthase 11 [Jatropha curcas] |
| 15 |
Hb_000277_020 |
0.1158319318 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 16 |
Hb_045810_010 |
0.1233608846 |
- |
- |
PREDICTED: laccase-12-like [Jatropha curcas] |
| 17 |
Hb_001510_050 |
0.1265386291 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_011924_030 |
0.1293705461 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_004656_030 |
0.1304556935 |
- |
- |
putative pectin methylesterase family protein [Populus trichocarpa] |
| 20 |
Hb_000035_360 |
0.1311586021 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |