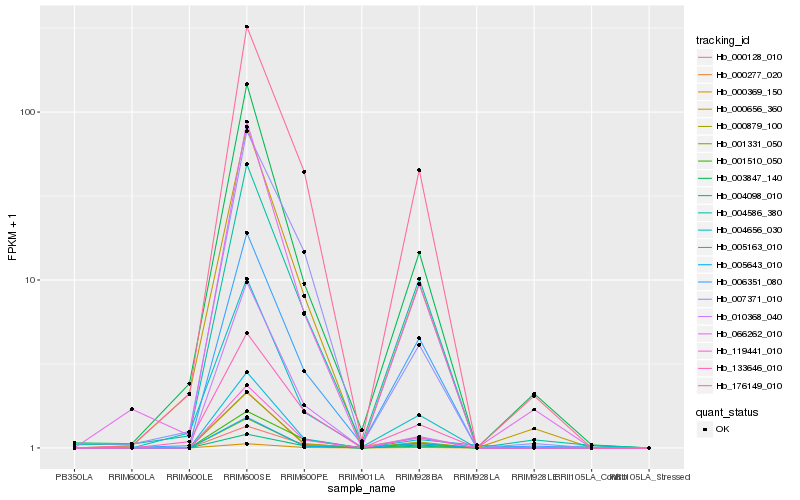
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000369_150 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20185 [Jatropha curcas] |
| 2 |
Hb_119441_010 |
0.0239272987 |
- |
- |
hypothetical protein CISIN_1g0477971mg, partial [Citrus sinensis] |
| 3 |
Hb_004098_010 |
0.0462002139 |
- |
- |
- |
| 4 |
Hb_000879_100 |
0.0685128899 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 5 |
Hb_000128_010 |
0.0723130637 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 6 |
Hb_176149_010 |
0.0731172476 |
- |
- |
hypothetical protein JCGZ_16793 [Jatropha curcas] |
| 7 |
Hb_005643_010 |
0.0735069686 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001331_050 |
0.0769096286 |
- |
- |
hypothetical protein VITISV_022439 [Vitis vinifera] |
| 9 |
Hb_000656_360 |
0.0878139936 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 10 |
Hb_006351_080 |
0.1012224419 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 11 |
Hb_066262_010 |
0.1019310146 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 12 |
Hb_001510_050 |
0.1029918484 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_004586_380 |
0.1065833881 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 14 |
Hb_003847_140 |
0.1085749023 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005163_010 |
0.1096455548 |
- |
- |
PREDICTED: probable terpene synthase 11 [Jatropha curcas] |
| 16 |
Hb_133646_010 |
0.1184681794 |
- |
- |
hypothetical protein RCOM_1044030 [Ricinus communis] |
| 17 |
Hb_004656_030 |
0.1293747029 |
- |
- |
putative pectin methylesterase family protein [Populus trichocarpa] |
| 18 |
Hb_007371_010 |
0.1311715373 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 19 |
Hb_000277_020 |
0.1312272691 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 20 |
Hb_010368_040 |
0.1331907741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |