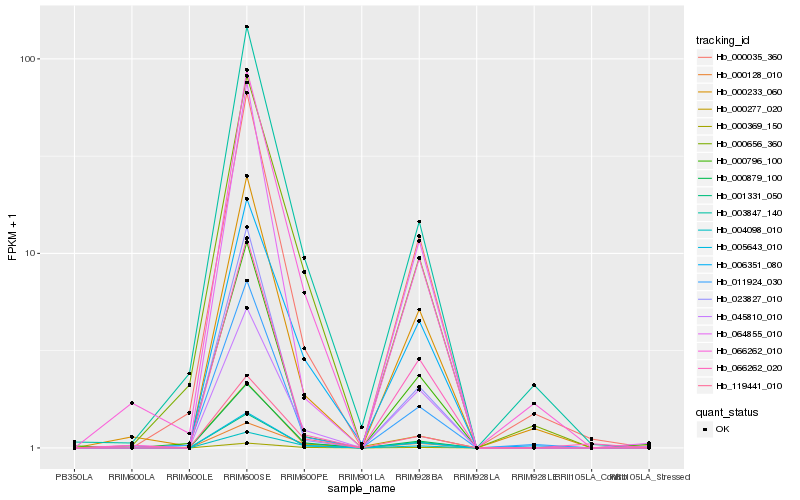
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005643_010 |
0.0 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001331_050 |
0.0222711262 |
- |
- |
hypothetical protein VITISV_022439 [Vitis vinifera] |
| 3 |
Hb_119441_010 |
0.0505183645 |
- |
- |
hypothetical protein CISIN_1g0477971mg, partial [Citrus sinensis] |
| 4 |
Hb_000879_100 |
0.0573296518 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 5 |
Hb_000369_150 |
0.0735069686 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20185 [Jatropha curcas] |
| 6 |
Hb_066262_010 |
0.0939561778 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 7 |
Hb_000277_020 |
0.0943299368 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 8 |
Hb_011924_030 |
0.0958096681 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_064855_010 |
0.0983085236 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_003847_140 |
0.0996017689 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000035_360 |
0.1024104774 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 12 |
Hb_000656_360 |
0.1031727737 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 13 |
Hb_045810_010 |
0.1039947242 |
- |
- |
PREDICTED: laccase-12-like [Jatropha curcas] |
| 14 |
Hb_000233_060 |
0.1045070798 |
- |
- |
hypothetical protein JCGZ_14793 [Jatropha curcas] |
| 15 |
Hb_006351_080 |
0.1057424579 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 16 |
Hb_066262_020 |
0.1079637174 |
- |
- |
hypothetical protein JCGZ_14803 [Jatropha curcas] |
| 17 |
Hb_000796_100 |
0.1095658945 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 18 |
Hb_004098_010 |
0.1110674955 |
- |
- |
- |
| 19 |
Hb_023827_010 |
0.1125203193 |
- |
- |
Cucumber peeling cupredoxin, putative [Ricinus communis] |
| 20 |
Hb_000128_010 |
0.1149764318 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |