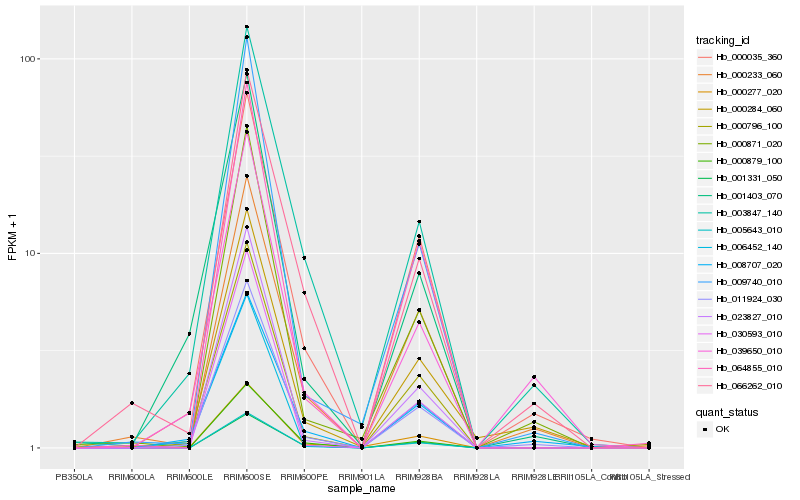
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011924_030 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000871_020 |
0.075696127 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_000035_360 |
0.0791112575 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 4 |
Hb_000284_060 |
0.0804306414 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 5 |
Hb_003847_140 |
0.0843227068 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000796_100 |
0.0864504689 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_009740_010 |
0.0867145772 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 8 |
Hb_008707_020 |
0.0875441517 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_039650_010 |
0.0875470903 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 10 |
Hb_023827_010 |
0.0900801145 |
- |
- |
Cucumber peeling cupredoxin, putative [Ricinus communis] |
| 11 |
Hb_064855_010 |
0.0955438875 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_005643_010 |
0.0958096681 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_066262_010 |
0.0991034154 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 14 |
Hb_001403_070 |
0.1003581284 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000233_060 |
0.1006798926 |
- |
- |
hypothetical protein JCGZ_14793 [Jatropha curcas] |
| 16 |
Hb_030593_010 |
0.104685983 |
- |
- |
hypothetical protein JCGZ_06063 [Jatropha curcas] |
| 17 |
Hb_006452_140 |
0.1059505745 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 18 |
Hb_000879_100 |
0.1061791552 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 19 |
Hb_001331_050 |
0.1064343879 |
- |
- |
hypothetical protein VITISV_022439 [Vitis vinifera] |
| 20 |
Hb_000277_020 |
0.1075583773 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |