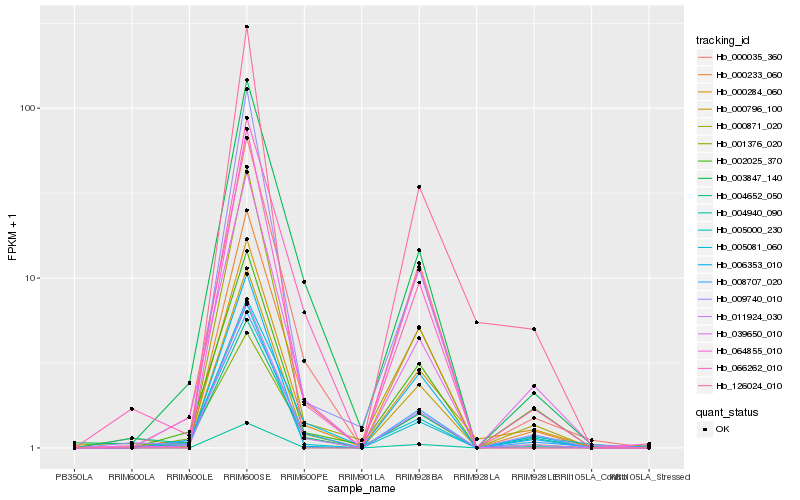
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 2 |
Hb_011924_030 |
0.0804306414 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_039650_010 |
0.0899986604 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 4 |
Hb_000035_360 |
0.0935512508 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 5 |
Hb_008707_020 |
0.0958742315 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000871_020 |
0.0987489733 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_000233_060 |
0.1040270949 |
- |
- |
hypothetical protein JCGZ_14793 [Jatropha curcas] |
| 8 |
Hb_003847_140 |
0.1076735014 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004940_090 |
0.1128164713 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 10 |
Hb_002025_370 |
0.1172926482 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_066262_010 |
0.1180007409 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 12 |
Hb_126024_010 |
0.1183378153 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Populus euphratica] |
| 13 |
Hb_006353_010 |
0.1202813584 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 14 |
Hb_005000_230 |
0.1225197901 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 15 |
Hb_009740_010 |
0.1239402776 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_000796_100 |
0.1255960928 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_064855_010 |
0.1276443589 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_004652_050 |
0.130188612 |
- |
- |
hypothetical protein POPTR_0001s15360g [Populus trichocarpa] |
| 19 |
Hb_005081_060 |
0.1305729487 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 20 |
Hb_001376_020 |
0.1313349052 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |