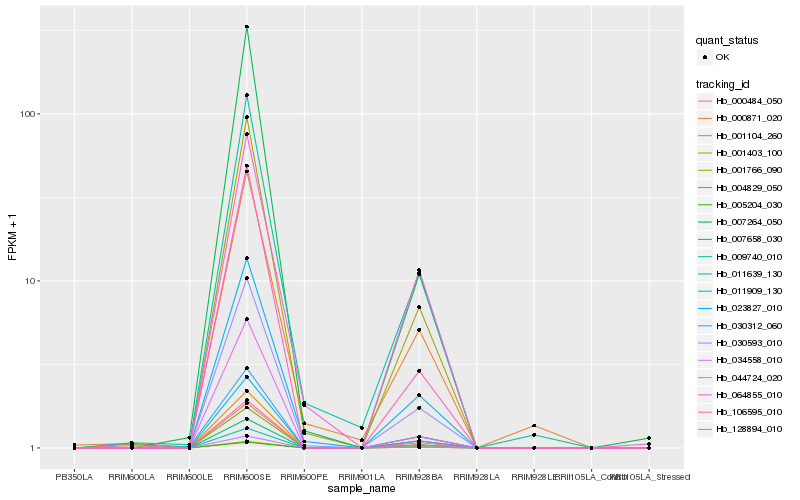
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030593_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06063 [Jatropha curcas] |
| 2 |
Hb_023827_010 |
0.0237388501 |
- |
- |
Cucumber peeling cupredoxin, putative [Ricinus communis] |
| 3 |
Hb_001766_090 |
0.0286424956 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 4 |
Hb_007658_030 |
0.0398733125 |
- |
- |
PREDICTED: lupeol synthase-like isoform X1 [Populus euphratica] |
| 5 |
Hb_000484_050 |
0.0413718874 |
- |
- |
hypothetical protein CICLE_v10028549mg [Citrus clementina] |
| 6 |
Hb_011909_130 |
0.0496604977 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 7 |
Hb_009740_010 |
0.0513926609 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 8 |
Hb_034558_010 |
0.0522967226 |
- |
- |
- |
| 9 |
Hb_128894_010 |
0.0636804016 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_106595_010 |
0.0761216505 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 11 |
Hb_011639_130 |
0.0773230367 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_004829_050 |
0.0785155959 |
- |
- |
hypothetical protein TRIUR3_09654 [Triticum urartu] |
| 13 |
Hb_064855_010 |
0.0847524217 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000871_020 |
0.085155178 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_001403_100 |
0.0852635572 |
- |
- |
hypothetical protein CICLE_v10017457mg, partial [Citrus clementina] |
| 16 |
Hb_001104_260 |
0.0856576368 |
- |
- |
hypothetical protein JCGZ_23628 [Jatropha curcas] |
| 17 |
Hb_044724_020 |
0.0862760811 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_030312_060 |
0.0865460165 |
- |
- |
PREDICTED: uncharacterized protein LOC105634226 [Jatropha curcas] |
| 19 |
Hb_007264_050 |
0.0910401731 |
- |
- |
- |
| 20 |
Hb_005204_030 |
0.094608059 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |