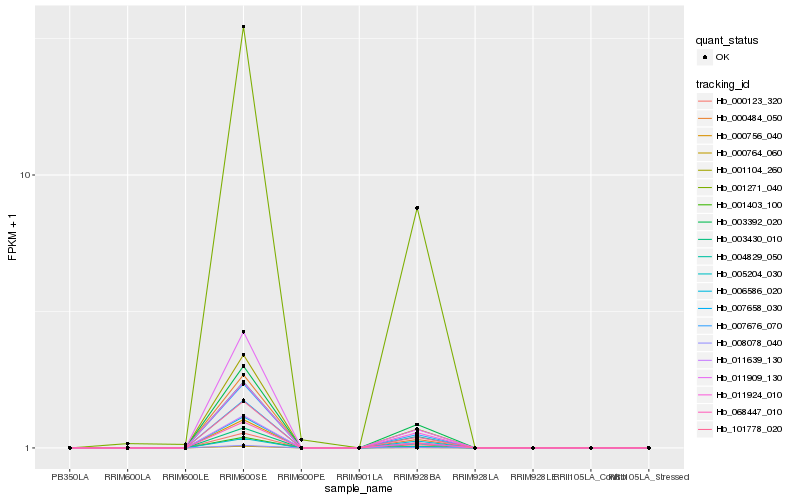
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004829_050 |
0.0 |
- |
- |
hypothetical protein TRIUR3_09654 [Triticum urartu] |
| 2 |
Hb_011639_130 |
0.0013956798 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_001403_100 |
0.0077788725 |
- |
- |
hypothetical protein CICLE_v10017457mg, partial [Citrus clementina] |
| 4 |
Hb_001104_260 |
0.00822774 |
- |
- |
hypothetical protein JCGZ_23628 [Jatropha curcas] |
| 5 |
Hb_005204_030 |
0.0182965706 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 6 |
Hb_008078_040 |
0.0224872166 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 7 |
Hb_000756_040 |
0.0238539944 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 8 |
Hb_000764_060 |
0.0279289573 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 9 |
Hb_007676_070 |
0.0293945178 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Populus euphratica] |
| 10 |
Hb_068447_010 |
0.0330928399 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_011909_130 |
0.0379971633 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 12 |
Hb_011924_010 |
0.0457782339 |
- |
- |
hypothetical protein, partial [Phaeodactylibacter xiamenensis] |
| 13 |
Hb_101778_020 |
0.0526545702 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000484_050 |
0.0559582922 |
- |
- |
hypothetical protein CICLE_v10028549mg [Citrus clementina] |
| 15 |
Hb_006586_020 |
0.0569333117 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_007658_030 |
0.0632569073 |
- |
- |
PREDICTED: lupeol synthase-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000123_320 |
0.0642532014 |
- |
- |
PREDICTED: MADS-box protein FLOWERING LOCUS C-like [Jatropha curcas] |
| 18 |
Hb_003430_010 |
0.0657491235 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 33 precursor [Populus trichocarpa] |
| 19 |
Hb_001271_040 |
0.0665388117 |
- |
- |
hypothetical protein POPTR_0019s039903g, partial [Populus trichocarpa] |
| 20 |
Hb_003392_020 |
0.0722900491 |
- |
- |
PREDICTED: uncharacterized protein LOC105650609 [Jatropha curcas] |