| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
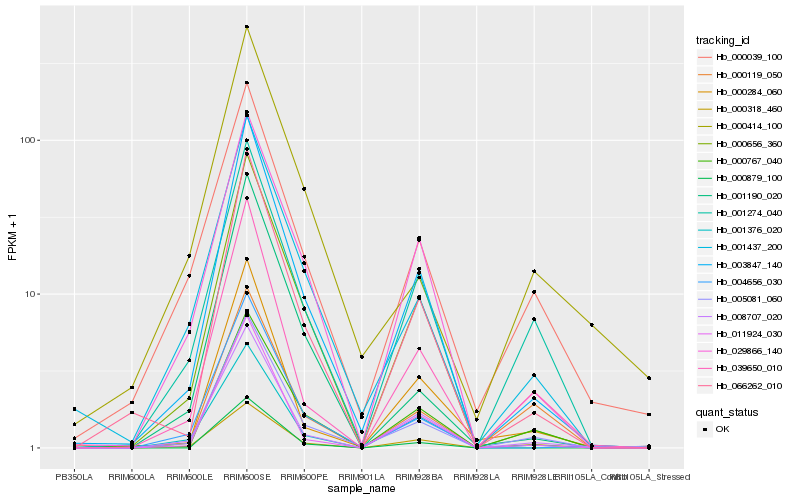
| 1 |
Hb_005081_060 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 2 |
Hb_003847_140 |
0.0818486925 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 3 |
Hb_039650_010 |
0.1024470712 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 4 |
Hb_000656_360 |
0.1090302963 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 5 |
Hb_001274_040 |
0.1092573882 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 6 |
Hb_008707_020 |
0.1106617627 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_001437_200 |
0.1110240406 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 8 |
Hb_000767_040 |
0.1143567783 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_066262_010 |
0.1144664379 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 10 |
Hb_000039_100 |
0.1188856457 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 11 |
Hb_000318_460 |
0.119765931 |
- |
- |
hypothetical protein CICLE_v10028333mg [Citrus clementina] |
| 12 |
Hb_011924_030 |
0.1249930457 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_029866_140 |
0.1286180447 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 14 |
Hb_000119_050 |
0.1290184471 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 15 |
Hb_000284_060 |
0.1305729487 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 16 |
Hb_004656_030 |
0.1309114872 |
- |
- |
putative pectin methylesterase family protein [Populus trichocarpa] |
| 17 |
Hb_001376_020 |
0.1326583442 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 18 |
Hb_000414_100 |
0.1401523214 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 19 |
Hb_000879_100 |
0.1411320058 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 20 |
Hb_001190_020 |
0.1426788914 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |