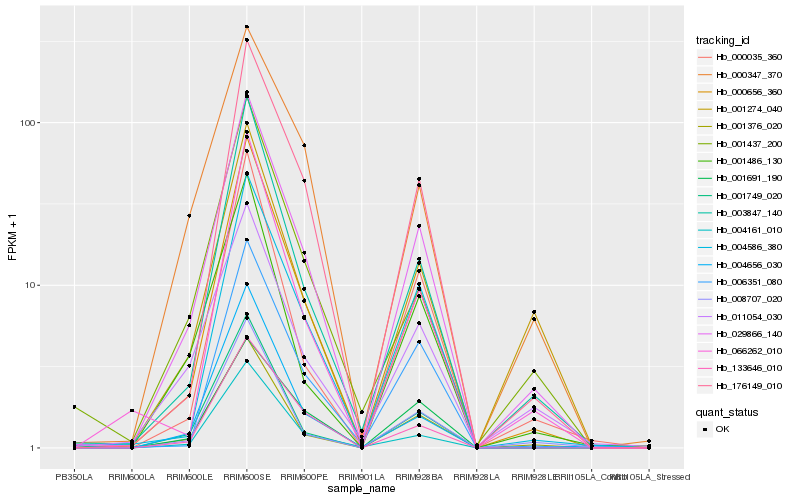
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029866_140 |
0.0 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 2 |
Hb_000656_360 |
0.0638210618 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 3 |
Hb_001376_020 |
0.0641085085 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 4 |
Hb_003847_140 |
0.0937566721 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008707_020 |
0.0955621774 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_176149_010 |
0.0977812844 |
- |
- |
hypothetical protein JCGZ_16793 [Jatropha curcas] |
| 7 |
Hb_011054_030 |
0.0995083145 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 8 |
Hb_001749_020 |
0.1015520231 |
- |
- |
hypothetical protein CISIN_1g042220mg, partial [Citrus sinensis] |
| 9 |
Hb_004586_380 |
0.1018447389 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 10 |
Hb_133646_010 |
0.109891642 |
- |
- |
hypothetical protein RCOM_1044030 [Ricinus communis] |
| 11 |
Hb_000347_370 |
0.1125286141 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 12 |
Hb_001486_130 |
0.1142501027 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 13 |
Hb_006351_080 |
0.1143720112 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 14 |
Hb_001274_040 |
0.1170140177 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 15 |
Hb_001437_200 |
0.1178928635 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 16 |
Hb_000035_360 |
0.120617483 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 17 |
Hb_001691_190 |
0.1217636404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004656_030 |
0.1244677403 |
- |
- |
putative pectin methylesterase family protein [Populus trichocarpa] |
| 19 |
Hb_004161_010 |
0.1248184306 |
- |
- |
hypothetical protein JCGZ_21278 [Jatropha curcas] |
| 20 |
Hb_066262_010 |
0.1259613208 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |