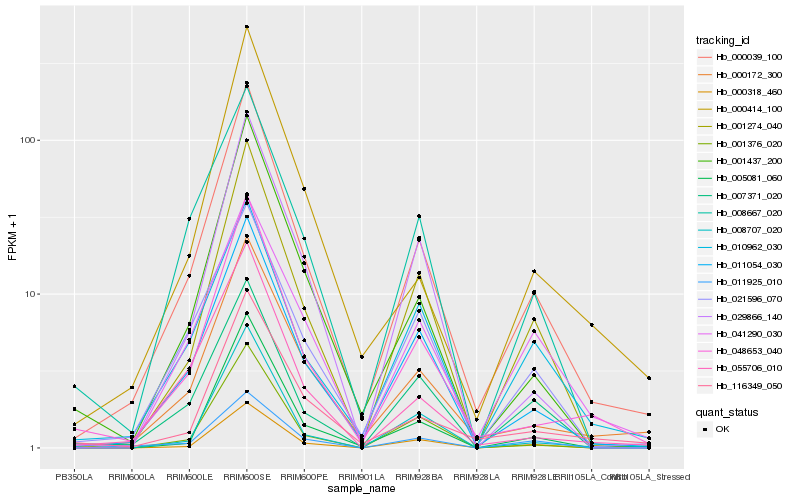
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000039_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 2 |
Hb_011054_030 |
0.0857799082 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 3 |
Hb_000172_300 |
0.0919805277 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 4 |
Hb_001274_040 |
0.0973632293 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 5 |
Hb_048653_040 |
0.1004829759 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 6 |
Hb_001437_200 |
0.1130552151 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 7 |
Hb_005081_060 |
0.1188856457 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 8 |
Hb_011925_010 |
0.1246420492 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_116349_050 |
0.1247127739 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 10 |
Hb_008707_020 |
0.1272619665 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_055706_010 |
0.1286497001 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 12 |
Hb_007371_020 |
0.12918471 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_021596_070 |
0.1300411442 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_008667_020 |
0.1302881831 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_010962_030 |
0.1302920582 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000414_100 |
0.1326059957 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 17 |
Hb_001376_020 |
0.1342531935 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 18 |
Hb_029866_140 |
0.1347668257 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 19 |
Hb_000318_460 |
0.1350574943 |
- |
- |
hypothetical protein CICLE_v10028333mg [Citrus clementina] |
| 20 |
Hb_041290_030 |
0.1369349711 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |