| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
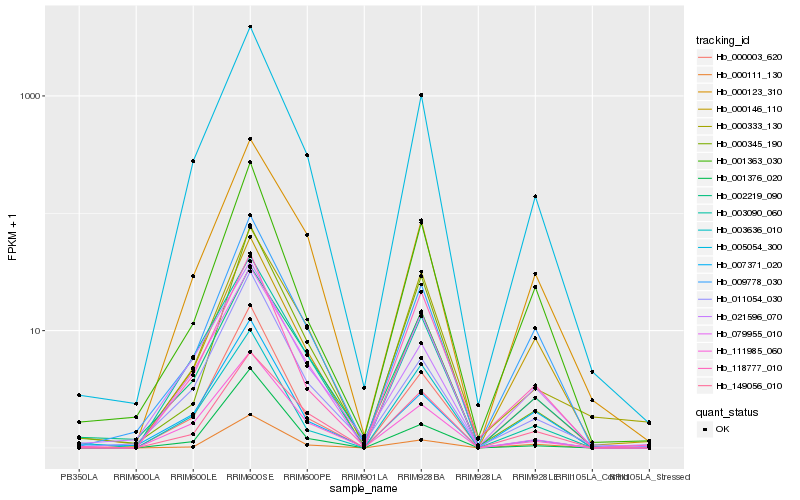
Hb_005054_300 |
0.0 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 2 |
Hb_111985_060 |
0.0731662565 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 3 |
Hb_011054_030 |
0.0885504188 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 4 |
Hb_003090_060 |
0.0922022784 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 5 |
Hb_009778_030 |
0.1013267715 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 6 |
Hb_000123_310 |
0.102461101 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 7 |
Hb_002219_090 |
0.1073896044 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 8 |
Hb_001363_030 |
0.109601044 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 9 |
Hb_001376_020 |
0.1110722199 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 10 |
Hb_021596_070 |
0.1141637355 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_000003_620 |
0.1147944859 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 12 |
Hb_149056_010 |
0.1155212594 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 13 |
Hb_079955_010 |
0.1186640452 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 14 |
Hb_118777_010 |
0.1207385904 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000146_110 |
0.121162807 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 16 |
Hb_007371_020 |
0.1219311453 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000333_130 |
0.1226244015 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000345_190 |
0.1252465865 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 19 |
Hb_003636_010 |
0.1253594535 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 20 |
Hb_000111_130 |
0.1259794403 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |