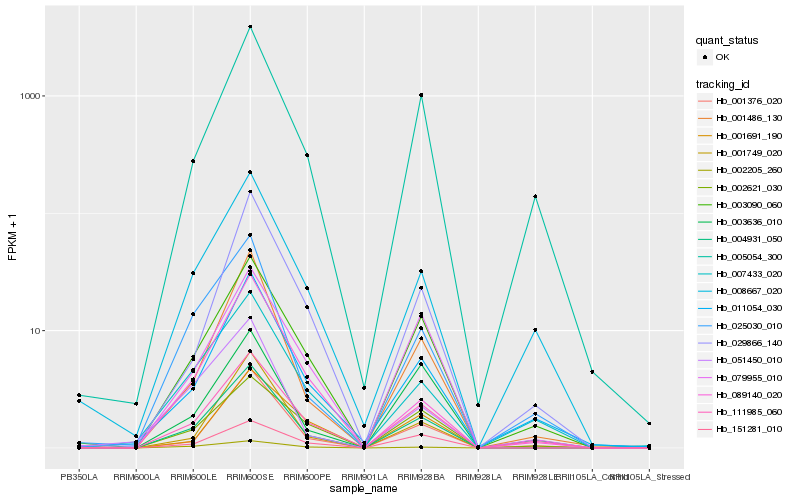
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004931_050 |
0.0 |
transcription factor |
TF Family: MYB-related |
Transcription repressor MYB4, putative [Ricinus communis] |
| 2 |
Hb_001486_130 |
0.1064894388 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 3 |
Hb_003090_060 |
0.1067672429 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 4 |
Hb_111985_060 |
0.1152705424 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 5 |
Hb_001749_020 |
0.1246935075 |
- |
- |
hypothetical protein CISIN_1g042220mg, partial [Citrus sinensis] |
| 6 |
Hb_079955_010 |
0.1286062258 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 7 |
Hb_001376_020 |
0.1300434311 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 8 |
Hb_011054_030 |
0.1330438952 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 9 |
Hb_005054_300 |
0.1355994309 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 10 |
Hb_025030_010 |
0.1361359587 |
- |
- |
- |
| 11 |
Hb_151281_010 |
0.1383655216 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 12 |
Hb_002205_260 |
0.1416622534 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 13 |
Hb_029866_140 |
0.1438681025 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 14 |
Hb_008667_020 |
0.1518819636 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_002621_030 |
0.1559186809 |
- |
- |
hypothetical protein CISIN_1g006155mg [Citrus sinensis] |
| 16 |
Hb_007433_020 |
0.1569159514 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_051450_010 |
0.1597361117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001691_190 |
0.1616358712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_089140_020 |
0.1637327888 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_003636_010 |
0.1675757598 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |