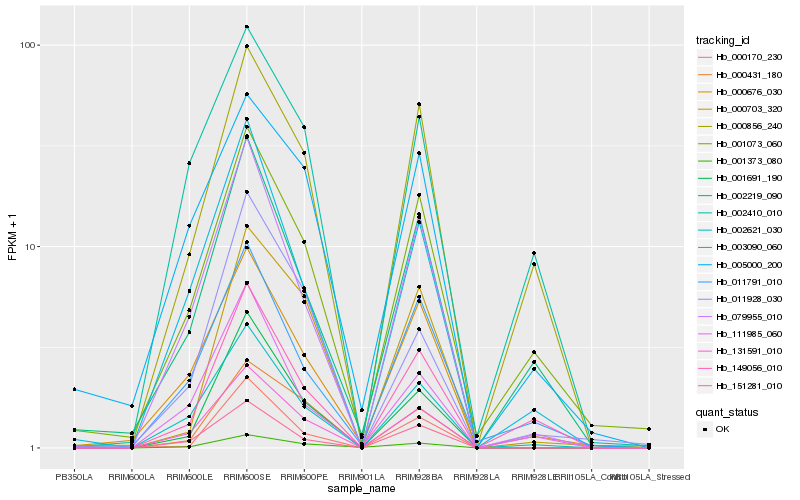
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_131591_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 2 |
Hb_151281_010 |
0.0806982445 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 3 |
Hb_002621_030 |
0.0844484422 |
- |
- |
hypothetical protein CISIN_1g006155mg [Citrus sinensis] |
| 4 |
Hb_079955_010 |
0.0981193888 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 5 |
Hb_001691_190 |
0.1099399344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000676_030 |
0.1183899264 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 7 |
Hb_003090_060 |
0.1187152664 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 8 |
Hb_000431_180 |
0.1197479182 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 9 |
Hb_001373_080 |
0.1239872099 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_011791_010 |
0.1487579386 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 11 |
Hb_111985_060 |
0.1498444129 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 12 |
Hb_001073_060 |
0.153843957 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 13 |
Hb_002410_010 |
0.1540706261 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 14 |
Hb_000856_240 |
0.155126415 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_005000_200 |
0.157762872 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |
| 16 |
Hb_002219_090 |
0.1606907831 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 17 |
Hb_011928_030 |
0.1614640888 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 18 |
Hb_000703_320 |
0.1638641682 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_000170_230 |
0.1640429497 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 20 |
Hb_149056_010 |
0.1646853653 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |