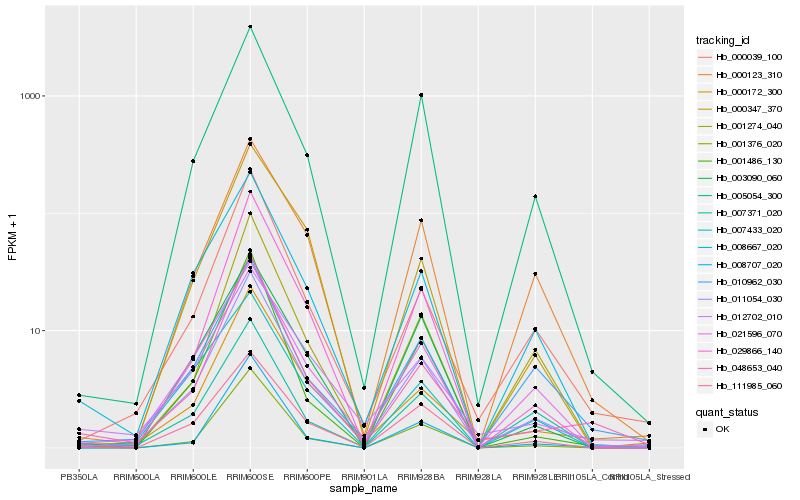
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011054_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 2 |
Hb_000039_100 |
0.0857799082 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 3 |
Hb_005054_300 |
0.0885504188 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 4 |
Hb_021596_070 |
0.0892574939 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_111985_060 |
0.0923723003 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 6 |
Hb_008667_020 |
0.0930257865 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_029866_140 |
0.0995083145 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 8 |
Hb_001376_020 |
0.1001015306 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 9 |
Hb_000123_310 |
0.1074269012 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 10 |
Hb_000172_300 |
0.1084273486 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 11 |
Hb_001274_040 |
0.1104178453 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 12 |
Hb_048653_040 |
0.1128890943 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 13 |
Hb_001486_130 |
0.1147130458 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 14 |
Hb_007371_020 |
0.1183456572 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_010962_030 |
0.1210590776 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_007433_020 |
0.121555093 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_008707_020 |
0.1244587202 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_003090_060 |
0.1272375859 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 19 |
Hb_000347_370 |
0.1284635158 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 20 |
Hb_012702_010 |
0.1300988848 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |