| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
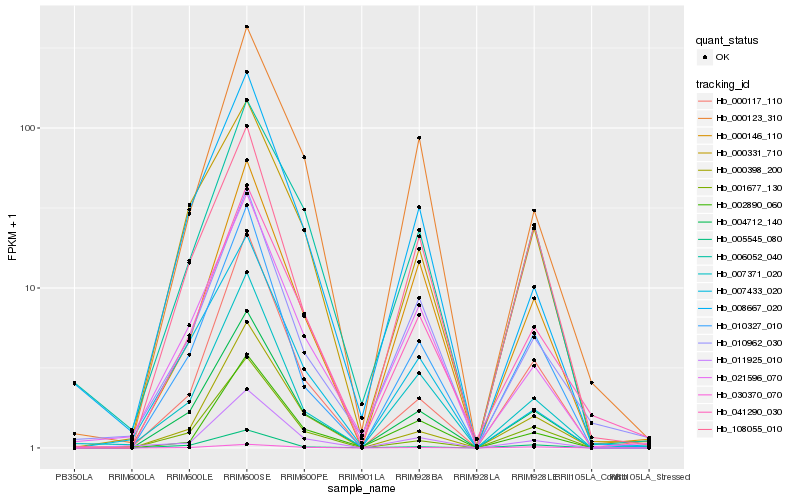
Hb_004712_140 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 2 |
Hb_041290_030 |
0.087365973 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 3 |
Hb_010327_010 |
0.0944256541 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_000331_710 |
0.1030442747 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_021596_070 |
0.106617166 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_000398_200 |
0.1078030083 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 7 |
Hb_001677_130 |
0.1085355451 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 8 |
Hb_007371_020 |
0.1094353232 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000117_110 |
0.1126829735 |
- |
- |
transferase, putative [Ricinus communis] |
| 10 |
Hb_010962_030 |
0.1145839556 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000146_110 |
0.1156675583 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 12 |
Hb_006052_040 |
0.1172961325 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 13 |
Hb_008667_020 |
0.1201394377 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_011925_010 |
0.1273141265 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000123_310 |
0.127389595 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 16 |
Hb_007433_020 |
0.131590371 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_005545_080 |
0.1330216161 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 18 |
Hb_002890_060 |
0.1332444305 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 19 |
Hb_108055_010 |
0.1337323802 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_030370_070 |
0.1379404873 |
- |
- |
ATP binding protein, putative [Ricinus communis] |