| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
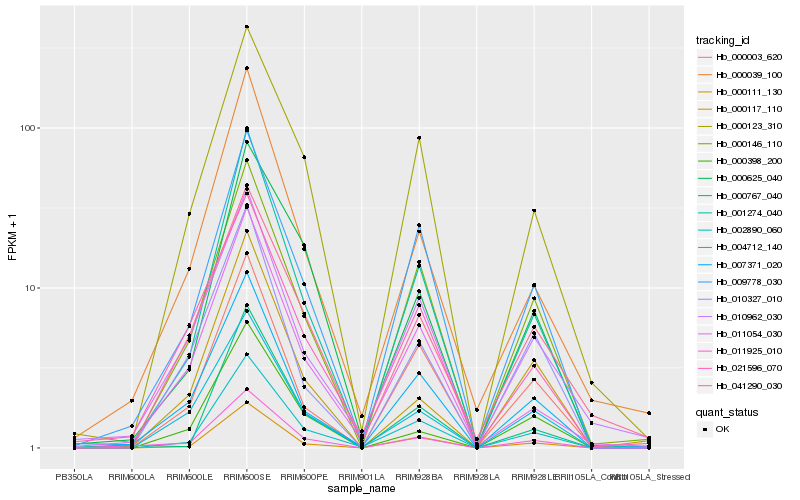
Hb_011925_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001274_040 |
0.1140949869 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 3 |
Hb_007371_020 |
0.1144194971 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002890_060 |
0.1203741919 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 5 |
Hb_009778_030 |
0.1222399713 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 6 |
Hb_000039_100 |
0.1246420492 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 7 |
Hb_000123_310 |
0.1262242892 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 8 |
Hb_004712_140 |
0.1273141265 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 9 |
Hb_000146_110 |
0.128815414 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 10 |
Hb_000398_200 |
0.1313370816 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 11 |
Hb_021596_070 |
0.134162353 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_000111_130 |
0.1374712826 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_000625_040 |
0.1389655273 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 14 |
Hb_011054_030 |
0.1392515731 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 15 |
Hb_010962_030 |
0.1394010072 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_041290_030 |
0.139830991 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 17 |
Hb_000117_110 |
0.1413813372 |
- |
- |
transferase, putative [Ricinus communis] |
| 18 |
Hb_010327_010 |
0.1431428755 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_000767_040 |
0.1463729116 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000003_620 |
0.147096968 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |