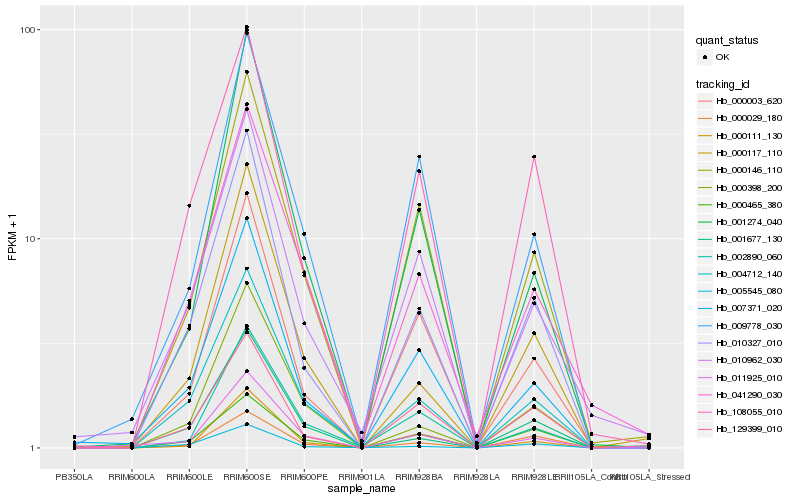
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010327_010 |
0.0 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_004712_140 |
0.0944256541 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 3 |
Hb_007371_020 |
0.0965353398 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_108055_010 |
0.1030635959 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000003_620 |
0.1125470207 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 6 |
Hb_000117_110 |
0.1192800688 |
- |
- |
transferase, putative [Ricinus communis] |
| 7 |
Hb_000146_110 |
0.1197959344 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 8 |
Hb_001677_130 |
0.1233049308 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 9 |
Hb_000029_180 |
0.1248308142 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 10 |
Hb_129399_010 |
0.1274738975 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_010962_030 |
0.129774153 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000398_200 |
0.1325310956 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 13 |
Hb_001274_040 |
0.1365903026 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 14 |
Hb_000465_380 |
0.1413717795 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_041290_030 |
0.1428340519 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 16 |
Hb_005545_080 |
0.142980451 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 17 |
Hb_011925_010 |
0.1431428755 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000111_130 |
0.1432801023 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 19 |
Hb_002890_060 |
0.1500985177 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 20 |
Hb_009778_030 |
0.1514299448 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |