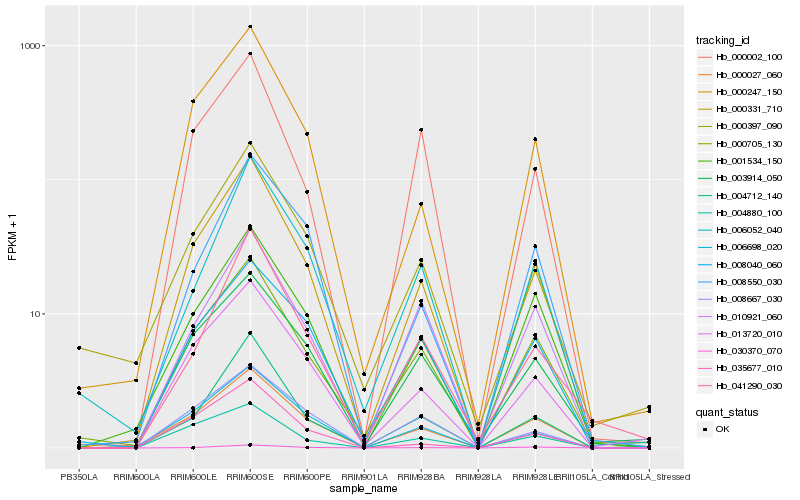
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_710 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_013720_010 |
0.0704717085 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 3 |
Hb_000027_060 |
0.0777769539 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 4 |
Hb_000247_150 |
0.0905781695 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_000705_130 |
0.094782548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004712_140 |
0.1030442747 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 7 |
Hb_006698_020 |
0.1041742782 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 8 |
Hb_001534_150 |
0.106623851 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 9 |
Hb_003914_050 |
0.110442291 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 10 |
Hb_004880_100 |
0.1209594179 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 11 |
Hb_035677_010 |
0.1303891736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_041290_030 |
0.1313784075 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 13 |
Hb_000397_090 |
0.1321176022 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 14 |
Hb_008667_030 |
0.1323226186 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 15 |
Hb_030370_070 |
0.1330829269 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000002_100 |
0.1333013973 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_008040_060 |
0.1334585724 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 18 |
Hb_006052_040 |
0.134736752 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 19 |
Hb_008550_030 |
0.1353318322 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 20 |
Hb_010921_060 |
0.1355506109 |
- |
- |
ATP binding protein, putative [Ricinus communis] |