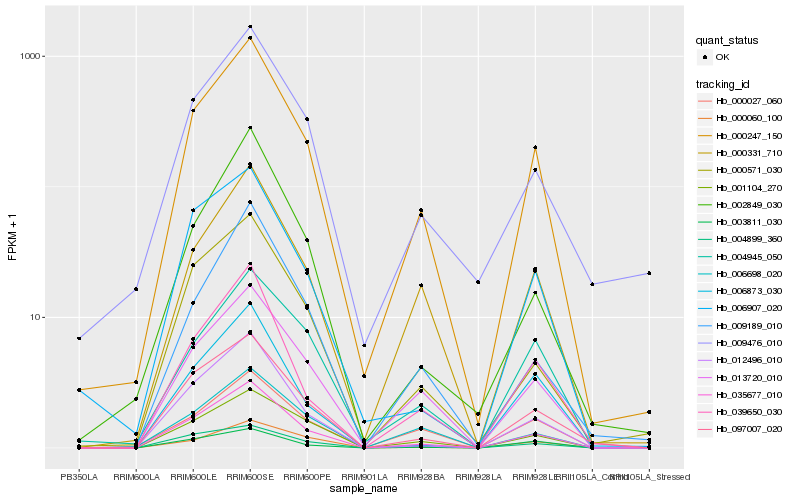
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000247_150 |
0.0 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 2 |
Hb_035677_010 |
0.0598842774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_013720_010 |
0.083009368 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 4 |
Hb_000331_710 |
0.0905781695 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_097007_020 |
0.1075639492 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 6 |
Hb_039650_030 |
0.1159051917 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 7 |
Hb_012496_010 |
0.116160505 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_001104_270 |
0.1166433007 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 9 |
Hb_000027_060 |
0.1198511579 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 10 |
Hb_009476_010 |
0.1219700447 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 11 |
Hb_004945_050 |
0.1224666007 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 12 |
Hb_000571_030 |
0.1231082048 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 13 |
Hb_004899_360 |
0.1289122645 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 14 |
Hb_006698_020 |
0.1314206102 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 15 |
Hb_009189_010 |
0.1317389289 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 16 |
Hb_006907_020 |
0.1352180498 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 17 |
Hb_003811_030 |
0.1358393822 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 18 |
Hb_000060_100 |
0.1364776178 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006873_030 |
0.1378772542 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 20 |
Hb_002849_030 |
0.1399029828 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |