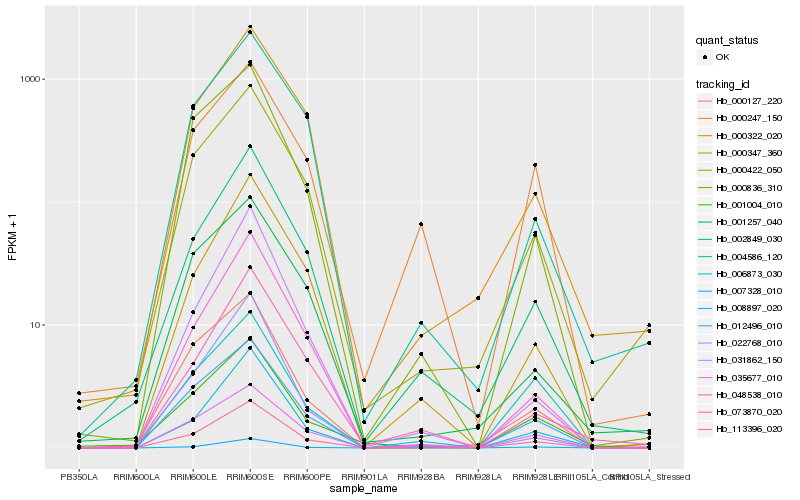
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113396_020 |
0.0 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 2 |
Hb_012496_010 |
0.0781914935 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_048538_010 |
0.0967582834 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 4 |
Hb_002849_030 |
0.1081844857 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 5 |
Hb_000422_050 |
0.1092346131 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 6 |
Hb_008897_020 |
0.1129920468 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 7 |
Hb_000127_220 |
0.1171020537 |
- |
- |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 8 |
Hb_000347_360 |
0.1179825516 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000322_020 |
0.118208494 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 10 |
Hb_001004_010 |
0.1183582327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_035677_010 |
0.1207454576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000836_310 |
0.1285274546 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 30 [Jatropha curcas] |
| 13 |
Hb_006873_030 |
0.1295805374 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_004586_120 |
0.1314602312 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 15 |
Hb_007328_010 |
0.1326720183 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_031862_150 |
0.1422750563 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 17 |
Hb_073870_020 |
0.144556673 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 18 |
Hb_000247_150 |
0.1460966762 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_001257_040 |
0.1475788545 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 20 |
Hb_022768_010 |
0.1502184794 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |