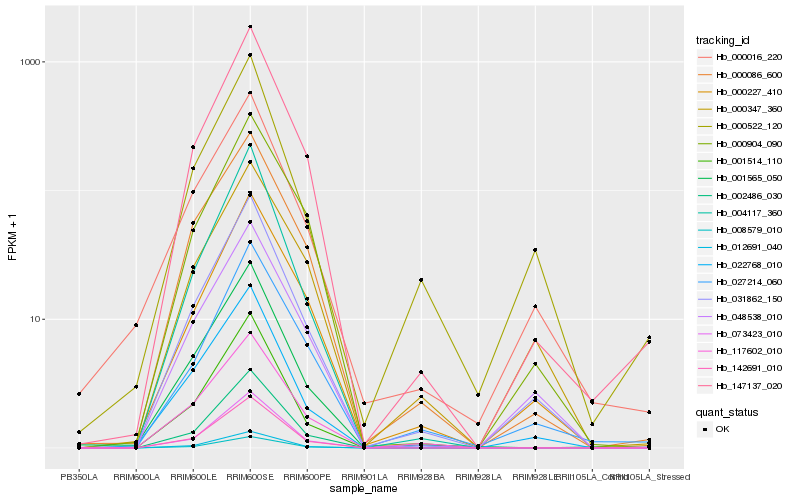
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_150 |
0.0 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 2 |
Hb_048538_010 |
0.0651298976 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 3 |
Hb_022768_010 |
0.0652363568 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_147137_020 |
0.0688093087 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 5 |
Hb_000227_410 |
0.0711099207 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 6 |
Hb_000086_600 |
0.0897834543 |
- |
- |
- |
| 7 |
Hb_142691_010 |
0.0911718672 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 8 |
Hb_000904_090 |
0.0923523701 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 9 |
Hb_008579_010 |
0.0926674033 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 10 |
Hb_000347_360 |
0.0926949969 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_012691_040 |
0.0940430919 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Fragaria vesca subsp. vesca] |
| 12 |
Hb_002486_030 |
0.0943950618 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 13 |
Hb_027214_060 |
0.0945701732 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 14 |
Hb_073423_010 |
0.0961560874 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000016_220 |
0.096673432 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 16 |
Hb_001565_050 |
0.0974767428 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 17 |
Hb_000522_120 |
0.0975150732 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 18 |
Hb_004117_360 |
0.1008205945 |
- |
- |
hypothetical protein JCGZ_00367 [Jatropha curcas] |
| 19 |
Hb_117602_010 |
0.1019789379 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 20 |
Hb_001514_110 |
0.10205958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |