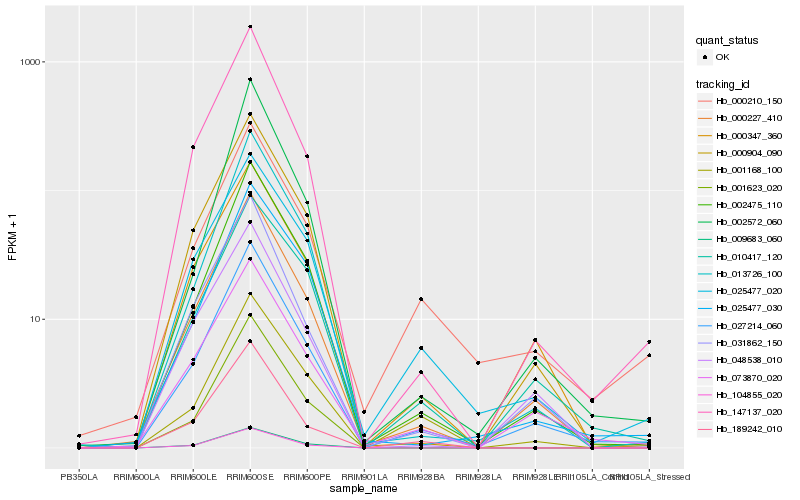
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027214_060 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 2 |
Hb_000227_410 |
0.0608251759 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 3 |
Hb_002475_110 |
0.0671588834 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 4 |
Hb_073870_020 |
0.0765793506 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 5 |
Hb_147137_020 |
0.0880110007 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 6 |
Hb_000904_090 |
0.0893278966 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 7 |
Hb_000347_360 |
0.0899367113 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031862_150 |
0.0945701732 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 9 |
Hb_013726_100 |
0.0964983887 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 10 |
Hb_010417_120 |
0.0981662981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_025477_020 |
0.1006804352 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 12 |
Hb_025477_030 |
0.1059613901 |
transcription factor |
TF Family: ERF |
dehydration-responsive element-binding protein 2 [Manihot esculenta] |
| 13 |
Hb_048538_010 |
0.1071175919 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 14 |
Hb_000210_150 |
0.1093771343 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_104855_020 |
0.1130913851 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002572_060 |
0.113512454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001623_020 |
0.1162245933 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 18 |
Hb_009683_060 |
0.1164160425 |
- |
- |
- |
| 19 |
Hb_001168_100 |
0.1177252554 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 20 |
Hb_189242_010 |
0.1180028483 |
- |
- |
hypothetical protein B456_013G0075002, partial [Gossypium raimondii] |