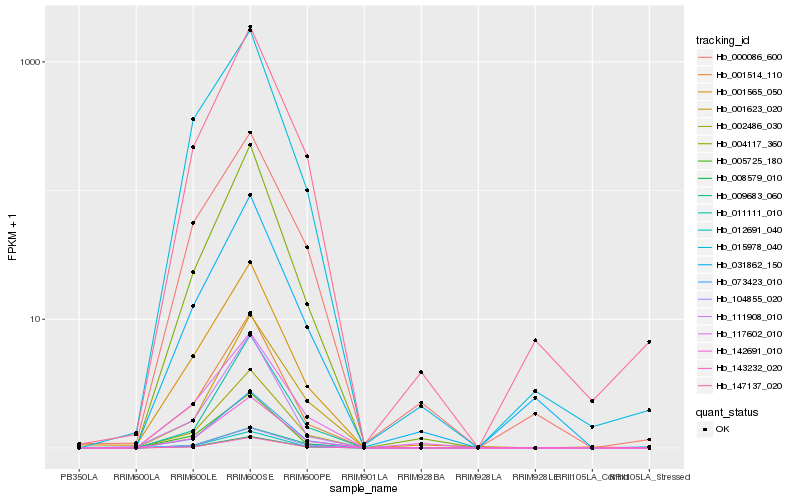
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142691_010 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 2 |
Hb_008579_010 |
0.0127748437 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 3 |
Hb_002486_030 |
0.0202082817 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 4 |
Hb_012691_040 |
0.0223908064 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Fragaria vesca subsp. vesca] |
| 5 |
Hb_073423_010 |
0.0262159707 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_001514_110 |
0.0449784517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_104855_020 |
0.0527792824 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_004117_360 |
0.0536475411 |
- |
- |
hypothetical protein JCGZ_00367 [Jatropha curcas] |
| 9 |
Hb_143232_020 |
0.0553355059 |
- |
- |
hypothetical protein JCGZ_20467 [Jatropha curcas] |
| 10 |
Hb_147137_020 |
0.0626691785 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 11 |
Hb_001565_050 |
0.0652160494 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 12 |
Hb_005725_180 |
0.0740855373 |
- |
- |
- |
| 13 |
Hb_117602_010 |
0.087348375 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_011111_010 |
0.087383955 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21A [Jatropha curcas] |
| 15 |
Hb_015978_040 |
0.0893836378 |
- |
- |
PREDICTED: uncharacterized protein LOC105642609 [Jatropha curcas] |
| 16 |
Hb_111908_010 |
0.0910379423 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_031862_150 |
0.0911718672 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 18 |
Hb_009683_060 |
0.0917241045 |
- |
- |
- |
| 19 |
Hb_000086_600 |
0.0948434295 |
- |
- |
- |
| 20 |
Hb_001623_020 |
0.0958458652 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |