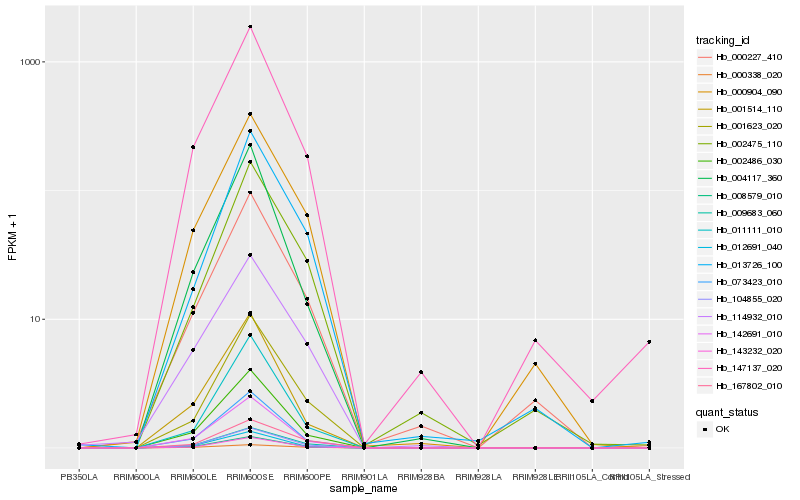
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104855_020 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_009683_060 |
0.042611794 |
- |
- |
- |
| 3 |
Hb_008579_010 |
0.0448555611 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 4 |
Hb_002486_030 |
0.0461010925 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 5 |
Hb_143232_020 |
0.0497423836 |
- |
- |
hypothetical protein JCGZ_20467 [Jatropha curcas] |
| 6 |
Hb_142691_010 |
0.0527792824 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 7 |
Hb_001623_020 |
0.0531968455 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 8 |
Hb_073423_010 |
0.0538960824 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_147137_020 |
0.067722271 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 10 |
Hb_000904_090 |
0.0721849646 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 11 |
Hb_012691_040 |
0.073022718 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Fragaria vesca subsp. vesca] |
| 12 |
Hb_167802_010 |
0.0783310124 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_114932_010 |
0.0832526994 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_013726_100 |
0.0882509698 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 15 |
Hb_011111_010 |
0.0898087824 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21A [Jatropha curcas] |
| 16 |
Hb_004117_360 |
0.0917202449 |
- |
- |
hypothetical protein JCGZ_00367 [Jatropha curcas] |
| 17 |
Hb_000227_410 |
0.0933278271 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 18 |
Hb_001514_110 |
0.0934035244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002475_110 |
0.0939781088 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 20 |
Hb_000338_020 |
0.0959422149 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |