| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
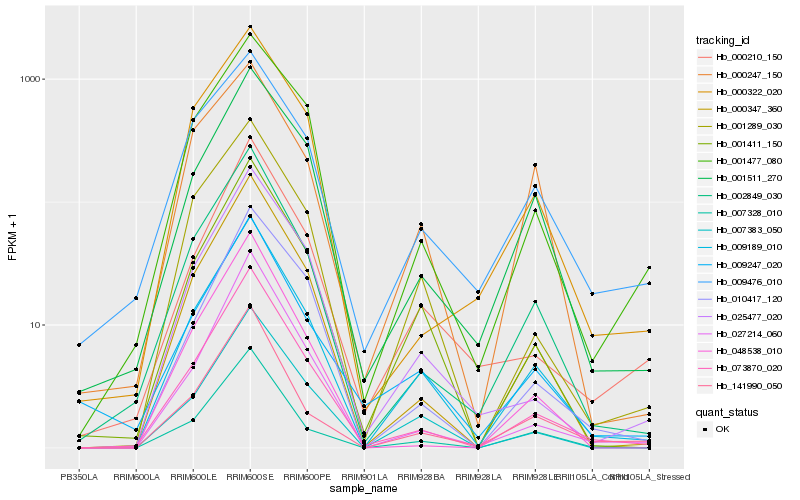
Hb_009189_010 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 2 |
Hb_001411_150 |
0.0766895106 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 3 |
Hb_073870_020 |
0.0778455932 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 4 |
Hb_007383_050 |
0.0824805869 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 5 |
Hb_002849_030 |
0.0876324818 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 6 |
Hb_000347_360 |
0.0904571084 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001289_030 |
0.093492324 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 8 |
Hb_007328_010 |
0.0978861989 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_001511_270 |
0.1110524873 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 10 |
Hb_001477_080 |
0.1125751574 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 11 |
Hb_141990_050 |
0.1126916791 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 12 |
Hb_009247_020 |
0.1191650858 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 13 |
Hb_025477_020 |
0.1196686886 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 14 |
Hb_027214_060 |
0.122614914 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 15 |
Hb_000210_150 |
0.123463218 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 16 |
Hb_000322_020 |
0.1248514775 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 17 |
Hb_010417_120 |
0.1262325371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009476_010 |
0.1307438309 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 19 |
Hb_000247_150 |
0.1317389289 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_048538_010 |
0.1318060365 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |