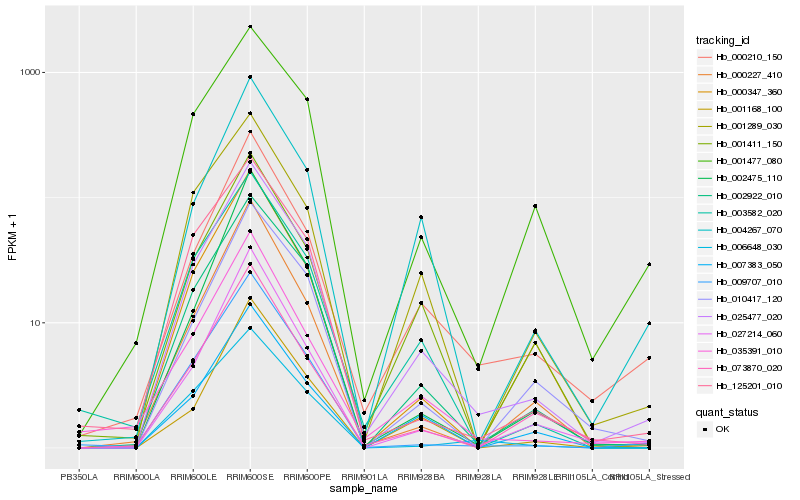
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025477_020 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 2 |
Hb_002922_010 |
0.0617868242 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_000210_150 |
0.0935773585 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 4 |
Hb_003582_020 |
0.0942088194 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 5 |
Hb_009707_010 |
0.0966192185 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 6 |
Hb_001289_030 |
0.0984011684 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 7 |
Hb_001477_080 |
0.098542705 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 8 |
Hb_027214_060 |
0.1006804352 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 9 |
Hb_010417_120 |
0.1012651753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001411_150 |
0.1042069048 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 11 |
Hb_035391_010 |
0.1042844018 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 12 |
Hb_073870_020 |
0.1048780205 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 13 |
Hb_000227_410 |
0.1055401139 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 14 |
Hb_000347_360 |
0.1069289058 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007383_050 |
0.1082411304 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 16 |
Hb_125201_010 |
0.1097077898 |
- |
- |
hypothetical protein POPTR_0004s03690g [Populus trichocarpa] |
| 17 |
Hb_004267_070 |
0.1099056641 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 18 |
Hb_001168_100 |
0.1134319085 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 19 |
Hb_006648_030 |
0.1143728487 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.3, putative [Ricinus communis] |
| 20 |
Hb_002475_110 |
0.1157622622 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |