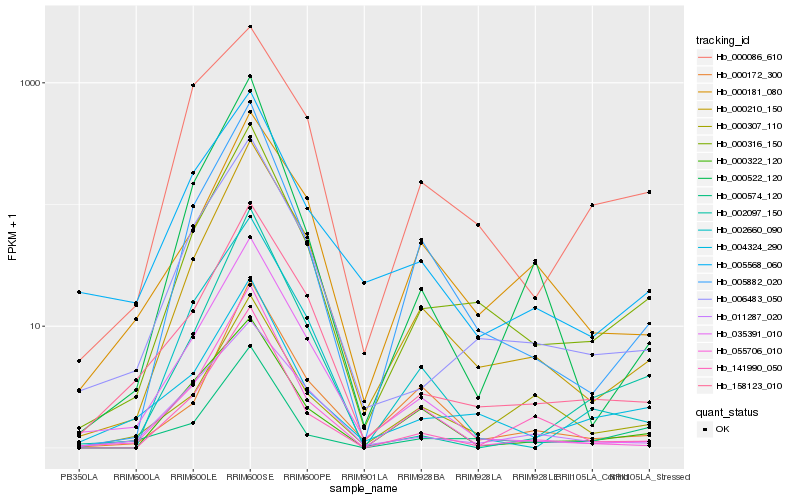
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000316_150 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_004324_290 |
0.1053528939 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 3 |
Hb_006483_050 |
0.1120820051 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 4 |
Hb_000210_150 |
0.1121285517 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 5 |
Hb_158123_010 |
0.1240982264 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_005882_020 |
0.1287212386 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 7 |
Hb_000574_120 |
0.1334231786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002097_150 |
0.1541593497 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 9 |
Hb_000086_610 |
0.1552794553 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 10 |
Hb_055706_010 |
0.1563926095 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 11 |
Hb_002660_090 |
0.156786893 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |
| 12 |
Hb_035391_010 |
0.1570108268 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 13 |
Hb_141990_050 |
0.1570926033 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 14 |
Hb_005568_060 |
0.1576194926 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 15 |
Hb_011287_020 |
0.1601046051 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000322_120 |
0.1608024127 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 17 |
Hb_000181_080 |
0.1644989496 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 18 |
Hb_000522_120 |
0.1650479777 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 19 |
Hb_000307_110 |
0.1663635732 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 20 |
Hb_000172_300 |
0.1682848857 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |