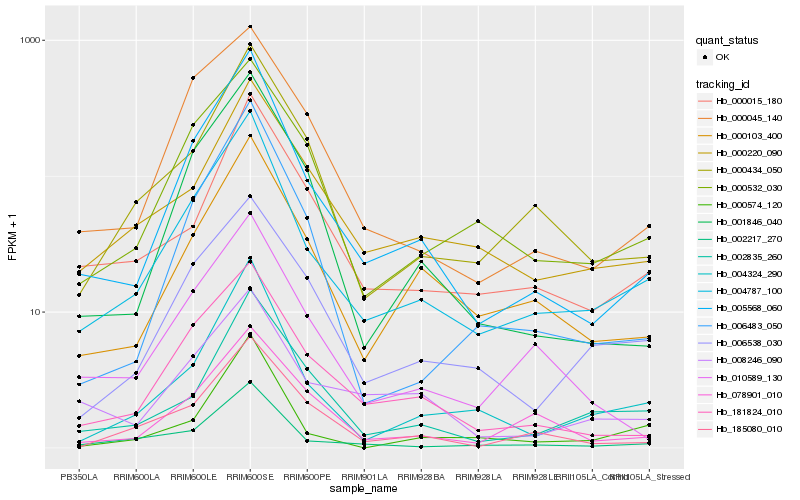
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004787_100 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 2 |
Hb_005568_060 |
0.1051821575 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 3 |
Hb_002217_270 |
0.1147465544 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 4 |
Hb_004324_290 |
0.1239263762 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 5 |
Hb_000532_030 |
0.1364543812 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 6 |
Hb_000103_400 |
0.1407595533 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006538_030 |
0.1409363838 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 8 |
Hb_181824_010 |
0.1418129552 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_000434_050 |
0.1423543535 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 10 |
Hb_000220_090 |
0.1550791936 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 11 |
Hb_000015_180 |
0.1561596926 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 12 |
Hb_000045_140 |
0.1577533965 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 13 |
Hb_002835_260 |
0.1608470728 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 14 |
Hb_010589_130 |
0.1609293544 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_185080_010 |
0.1642234829 |
- |
- |
- |
| 16 |
Hb_000574_120 |
0.1652428579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008246_090 |
0.1674085878 |
- |
- |
- |
| 18 |
Hb_006483_050 |
0.1677032461 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 19 |
Hb_001846_040 |
0.1694232208 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_078901_010 |
0.1709738244 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |