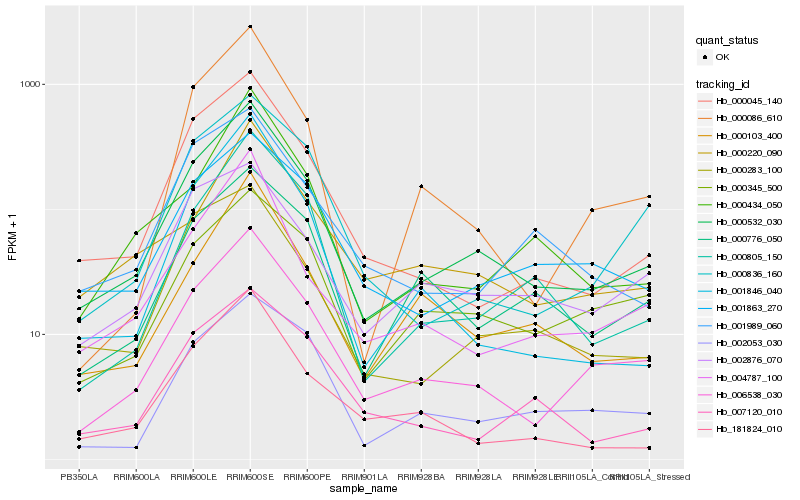
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000532_030 |
0.0 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 2 |
Hb_006538_030 |
0.092879266 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 3 |
Hb_000045_140 |
0.1154102509 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 4 |
Hb_000283_100 |
0.1253130768 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000805_150 |
0.1274761682 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_001863_270 |
0.1317917804 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 7 |
Hb_004787_100 |
0.1364543812 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 8 |
Hb_000434_050 |
0.1387957908 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 9 |
Hb_181824_010 |
0.1402624119 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_000103_400 |
0.1420419509 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001989_060 |
0.1425582537 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002053_030 |
0.1427791089 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000776_050 |
0.1440514545 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 14 |
Hb_001846_040 |
0.1449389906 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_007120_010 |
0.1466487862 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 16 |
Hb_000836_160 |
0.1475010773 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 17 |
Hb_000220_090 |
0.1485684477 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 18 |
Hb_000086_610 |
0.1488047588 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 19 |
Hb_000345_500 |
0.1497079408 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 20 |
Hb_002876_070 |
0.1508461396 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |