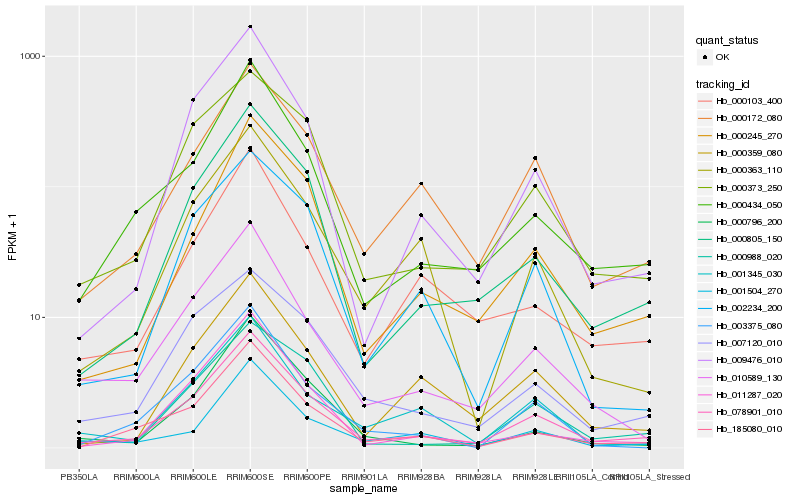
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078901_010 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 2 |
Hb_000805_150 |
0.0972525016 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000434_050 |
0.1051315107 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 4 |
Hb_009476_010 |
0.1110597126 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 5 |
Hb_000245_270 |
0.1131403329 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 6 |
Hb_000172_080 |
0.1177727918 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 7 |
Hb_000373_250 |
0.1181202021 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 8 |
Hb_010589_130 |
0.1201483968 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_185080_010 |
0.1212473755 |
- |
- |
- |
| 10 |
Hb_007120_010 |
0.1291480843 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 11 |
Hb_000988_020 |
0.1314040877 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000359_080 |
0.133817273 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 13 |
Hb_002234_200 |
0.134428628 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 14 |
Hb_011287_020 |
0.1387543799 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000103_400 |
0.1394160012 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003375_080 |
0.1420534479 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001504_270 |
0.1421638247 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 18 |
Hb_001345_030 |
0.1437646979 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 19 |
Hb_000363_110 |
0.144807271 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 20 |
Hb_000796_200 |
0.148708804 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |